MicroRNA-1 attenuates the growth and metastasis of small cell lung cancer through CXCR4/FOXM1/RRM2 axis
- PMID: 36597126
- PMCID: PMC9811802
- DOI: 10.1186/s12943-022-01695-6
MicroRNA-1 attenuates the growth and metastasis of small cell lung cancer through CXCR4/FOXM1/RRM2 axis
Abstract
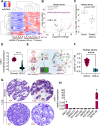
Background: Small cell lung cancer (SCLC) is an aggressive lung cancer subtype that is associated with high recurrence and poor prognosis. Due to lack of potential drug targets, SCLC patients have few therapeutic options. MicroRNAs (miRNAs) provide an interesting repertoire of therapeutic molecules; however, the identification of miRNAs regulating SCLC growth and metastasis and their precise regulatory mechanisms are not well understood.
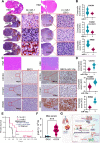
Methods: To identify novel miRNAs regulating SCLC, we performed miRNA-sequencing from donor/patient serum samples and analyzed the bulk RNA-sequencing data from the tumors of SCLC patients. Further, we developed a nanotechnology-based, highly sensitive method to detect microRNA-1 (miR-1, identified miRNA) in patient serum samples and SCLC cell lines. To assess the therapeutic potential of miR-1, we developed various in vitro models, including miR-1 sponge (miR-1Zip) and DOX-On-miR-1 (Tet-ON) inducible stable overexpression systems. Mouse models derived from intracardiac injection of SCLC cells (miR-1Zip and DOX-On-miR-1) were established to delineate the role of miR-1 in SCLC metastasis. In situ hybridization and immunohistochemistry were used to analyze the expression of miR-1 and target proteins (mouse and human tumor specimens), respectively. Dual-luciferase assay was used to validate the target of miR-1, and chromatin immunoprecipitation assay was used to investigate the protein-gene interactions.
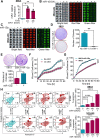
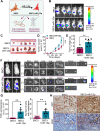
Results: A consistent downregulation of miR-1 was observed in tumor tissues and serum samples of SCLC patients compared to their matched normal controls, and these results were recapitulated in SCLC cell lines. Gain of function studies of miR-1 in SCLC cell lines showed decreased cell growth and oncogenic signaling, whereas loss of function studies of miR-1 rescued this effect. Intracardiac injection of gain of function of miR-1 SCLC cell lines in the mouse models showed a decrease in distant organ metastasis, whereas loss of function of miR-1 potentiated growth and metastasis. Mechanistic studies revealed that CXCR4 is a direct target of miR-1 in SCLC. Using unbiased transcriptomic analysis, we identified CXCR4/FOXM1/RRM2 as a unique axis that regulates SCLC growth and metastasis. Our results further showed that FOXM1 directly binds to the RRM2 promoter and regulates its activity in SCLC.
Conclusions: Our findings revealed that miR-1 is a critical regulator for decreasing SCLC growth and metastasis. It targets the CXCR4/FOXM1/RRM2 axis and has a high potential for the development of novel SCLC therapies. MicroRNA-1 (miR-1) downregulation in the tumor tissues and serum samples of SCLC patients is an important hallmark of tumor growth and metastasis. The introduction of miR-1 in SCLC cell lines decreases cell growth and metastasis. Mechanistically, miR-1 directly targets CXCR4, which further prevents FOXM1 binding to the RRM2 promoter and decreases SCLC growth and metastasis.
Keywords: CXCR4; FOXM1; Neuroendocrine carcinoma; RRM2; Small cell lung cancer; microRNAs.
© 2023. The Author(s).
Conflict of interest statement
SKB is co-founder of Sanguine Diagnostics and Therapeutics, Inc. AKG has served on Advisory Boards for AstraZeneca, Jazz pharmaceuticals, Sanofi–Genzyme, Beigene; received research support from Takeda Pharmaceuticals, Merck, TAB Biosciences, IOVANCE, and Mirati Therapeutics; is a consultant for Flagship Biosciences and serves on the DSMC for Y-mAbs Therapeutics. Other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

