Shedding light on structure, function and regulation of human sirtuins: a comprehensive review
- PMID: 36597461
- PMCID: PMC9805487
- DOI: 10.1007/s13205-022-03455-1
Shedding light on structure, function and regulation of human sirtuins: a comprehensive review
Abstract
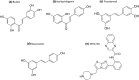
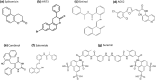
Sirtuins play an important role in signalling pathways associated with various metabolic regulations. They possess mono-ADP-ribosyltransferase or deacylase activity like demalonylase, deacetylase, depalmitoylase, demyristoylase and desuccinylase activity. Sirtuins are histone deacetylases which depends upon nicotinamide adenine dinucleotide (NAD) that deacetylate lysine residues. There are a total of seven human sirtuins that have been identified namely, SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6 and SIRT7. The subcellular location of mammalian sirtuins, SIRT1, SIRT6, and SIRT7 are in the nucleus; SIRT3, SIRT4, and SIRT5 are in mitochondria, and SIRT2 is in cytoplasm. Structurally sirtuins contains a N-terminal, a C-terminal and a Zn+ binding domain. The sirtuin family has been found to be crucial for maintaining lipid and glucose homeostasis, and also for regulating insulin secretion and sensitivity, DNA repair pathways, neurogenesis, inflammation, and ageing. Based on the literature, sirtuins are overexpressed and play an important role in tumorigenicity in various types of cancer such as non-small cell lung cancer, colorectal cancer, etc. In this review, we have discussed about the different types of human sirtuins along with their structural and functional features. We have also discussed about the various natural and synthetic regulators of sirtuin activities like resveratrol. Our overall study shows that the correct regulation of sirtuins can be a good target for preventing and treating various diseases for improving the human lifespan. To investigate the true therapeutic potential of sirtuin proteins and their efficacy in a variety of pathological diseases, a better knowledge of the link between the structure and function of sirtuin proteins would be necessary.
Keywords: Ageing; Deacetylase; Nicotinamide adenine dinucleotide; Rossmann fold; Sirtuins.
© King Abdulaziz City for Science and Technology 2022, Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest in the publication.
Figures


Similar articles
-
Screening Analysis of Sirtuins Family Expression on Anti-Inflammation of Resveratrol in Endothelial Cells.Med Sci Monit. 2019 Jun 3;25:4137-4148. doi: 10.12659/MSM.913240. Med Sci Monit. 2019. PMID: 31158122 Free PMC article.
-
Sirtuin family in autoimmune diseases.Front Immunol. 2023 Jul 6;14:1186231. doi: 10.3389/fimmu.2023.1186231. eCollection 2023. Front Immunol. 2023. PMID: 37483618 Free PMC article. Review.
-
The role of sirtuins in cardiac disease.Am J Physiol Heart Circ Physiol. 2015 Nov;309(9):H1375-89. doi: 10.1152/ajpheart.00053.2015. Epub 2015 Jul 31. Am J Physiol Heart Circ Physiol. 2015. PMID: 26232232 Free PMC article. Review.
-
Current understanding and future perspectives of the roles of sirtuins in the reprogramming and differentiation of pluripotent stem cells.Exp Biol Med (Maywood). 2018 Mar;243(6):563-575. doi: 10.1177/1535370218759636. Exp Biol Med (Maywood). 2018. PMID: 29557214 Free PMC article. Review.
-
Protective effects of sirtuins in cardiovascular diseases: from bench to bedside.Eur Heart J. 2015 Dec 21;36(48):3404-12. doi: 10.1093/eurheartj/ehv290. Epub 2015 Jun 25. Eur Heart J. 2015. PMID: 26112889 Free PMC article. Review.
Cited by
-
Age-Associated Changes of Sirtuin 2 Expression in CNS and the Periphery.Biology (Basel). 2023 Nov 29;12(12):1476. doi: 10.3390/biology12121476. Biology (Basel). 2023. PMID: 38132302 Free PMC article. Review.
-
Exploring Sirtuins: New Frontiers in Managing Heart Failure with Preserved Ejection Fraction.Int J Mol Sci. 2024 Jul 15;25(14):7740. doi: 10.3390/ijms25147740. Int J Mol Sci. 2024. PMID: 39062982 Free PMC article. Review.
-
SIRT3/6: an amazing challenge and opportunity in the fight against fibrosis and aging.Cell Mol Life Sci. 2024 Jan 31;81(1):69. doi: 10.1007/s00018-023-05093-z. Cell Mol Life Sci. 2024. PMID: 38294557 Free PMC article. Review.
-
Back to the Basics: Usefulness of Naturally Aged Mouse Models and Immunohistochemical and Quantitative Morphologic Methods in Studying Mechanisms of Lung Aging and Associated Diseases.Biomedicines. 2023 Jul 24;11(7):2075. doi: 10.3390/biomedicines11072075. Biomedicines. 2023. PMID: 37509714 Free PMC article. Review.
-
Symmetrical 2,7-disubstituted 9H-fluoren-9-one as a novel and promising scaffold for selective targeting of SIRT2.Arch Pharm (Weinheim). 2024 Dec;357(12):e2400661. doi: 10.1002/ardp.202400661. Epub 2024 Sep 28. Arch Pharm (Weinheim). 2024. PMID: 39340291 Free PMC article.
References
-
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani O, Li W, Chua KF. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487(7405):114–118. doi: 10.1038/nature11043. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

