This is a preprint.
A machine learning-based phenotype for long COVID in children: an EHR-based study from the RECOVER program
- PMID: 36597534
- PMCID: PMC9810222
- DOI: 10.1101/2022.12.22.22283791
A machine learning-based phenotype for long COVID in children: an EHR-based study from the RECOVER program
Update in
-
A machine learning-based phenotype for long COVID in children: An EHR-based study from the RECOVER program.PLoS One. 2023 Aug 10;18(8):e0289774. doi: 10.1371/journal.pone.0289774. eCollection 2023. PLoS One. 2023. PMID: 37561683 Free PMC article.
Abstract
Background: As clinical understanding of pediatric Post-Acute Sequelae of SARS CoV-2 (PASC) develops, and hence the clinical definition evolves, it is desirable to have a method to reliably identify patients who are likely to have post-acute sequelae of SARS CoV-2 (PASC) in health systems data.
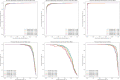
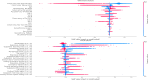
Methods and findings: In this study, we developed and validated a machine learning algorithm to classify which patients have PASC (distinguishing between Multisystem Inflammatory Syndrome in Children (MIS-C) and non-MIS-C variants) from a cohort of patients with positive SARS-CoV-2 test results in pediatric health systems within the PEDSnet EHR network. Patient features included in the model were selected from conditions, procedures, performance of diagnostic testing, and medications using a tree-based scan statistic approach. We used an XGboost model, with hyperparameters selected through cross-validated grid search, and model performance was assessed using 5-fold cross-validation. Model predictions and feature importance were evaluated using Shapley Additive exPlanation (SHAP) values.
Conclusions: The model provides a tool for identifying patients with PASC and an approach to characterizing PASC using diagnosis, medication, laboratory, and procedure features in health systems data. Using appropriate threshold settings, the model can be used to identify PASC patients in health systems data at higher precision for inclusion in studies or at higher recall in screening for clinical trials, especially in settings where PASC diagnosis codes are used less frequently or less reliably. Analysis of how specific features contribute to the classification process may assist in gaining a better understanding of features that are associated with PASC diagnoses.
Funding source: This research was funded by the National Institutes of Health (NIH) Agreement OT2HL161847-01 as part of the Researching COVID to Enhance Recovery (RECOVER) program of research.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the RECOVER Program, the NIH or other funders.
Figures
References
-
- Rao S, Lee GM, Razzaghi H, et al. Clinical features and burden of post-acute sequelae of SARS-CoV-2 infection in children and adolescents: an exploratory EHR-based cohort study from the RECOVER program. MedRxiv Prepr Serv Health Sci. Published online May 25, 2022:2022.05.24.22275544. doi: 10.1101/2022.05.24.22275544 - DOI
-
- Reese J, Blau H, Bergquist T, et al. Generalizable Long COVID Subtypes: Findings from the NIH N3C and RECOVER Programs. MedRxiv Prepr Serv Health Sci. Published online May 25, 2022:2022.05.24.22275398. doi: 10.1101/2022.05.24.22275398 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
