Modelling the Gastrointestinal Carriage of Klebsiella pneumoniae Infections
- PMID: 36598189
- PMCID: PMC9972987
- DOI: 10.1128/mbio.03121-22
Modelling the Gastrointestinal Carriage of Klebsiella pneumoniae Infections
Abstract
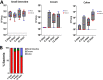
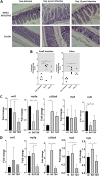
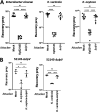
Klebsiella pneumoniae is a leading cause of nosocomial and community acquired infections, making K. pneumoniae the pathogen that is associated with the second largest number of deaths attributed to any antibiotic resistant infection. K. pneumoniae colonizes the nasopharynx and the gastrointestinal tract in an asymptomatic manner without dissemination to other tissues. Importantly, gastrointestinal colonization is a requisite for infection. Our understanding of K. pneumoniae colonization is still based on interrogating mouse models in which animals are pretreated with antibiotics to disturb the colonization resistance imposed by the gut microbiome. In these models, infections disseminate to other tissues. Here, we report a murine model to allow for the study of the gastrointestinal colonization of K. pneumoniae without tissue dissemination. Hypervirulent and antibiotic resistant strains stably colonize the gastrointestinal tract of in an inbred mouse population without antibiotic treatment. The small intestine is the primary site of colonization and is followed by a transition to the colon over time, without dissemination to other tissues. Our model recapitulates the disease dynamics of the metastatic K. pneumoniae strains that are able to disseminate from the gastrointestinal tract to other sterile sites. Colonization is associated with mild to moderate histopathology, no significant inflammation, and no effect on the richness of the microbiome. Our model sums up the clinical scenario in which antibiotic treatment disturbs the colonization of K. pneumoniae and results in dissemination to other tissues. Finally, we establish that the capsule polysaccharide is necessary for the colonization of the large intestine, whereas the type VI secretion system contributes to colonization across the gastrointestinal tract. IMPORTANCE Klebsiella pneumoniae is one of the pathogens that is sweeping the world in the antibiotic resistance pandemic. Klebsiella colonizes the nasopharynx and the gut of healthy subjects in an asymptomatic manner, making gut colonization a requisite for infection. This makes it essential to understand the gastrointestinal carriage in preventing Klebsiella infections. Current research models rely on the perturbation of the gut microbiome by antibiotics, resulting in an invasive infection. Here, we report a new model of K. pneumoniae gut colonization that recapitulates key features of the asymptomatic human gastrointestinal tract colonization. In our model, there is no need to disturb the microbiota to achieve stable colonization, and there is no dissemination to other tissues. Our model sums up the clinical scenario in which antibiotic treatment triggers invasive infection. We envision that our model will be an excellent platform upon which to investigate factors enhancing colonization and invasive infections and to test therapeutics to eliminate Klebsiella asymptomatic colonization.
Keywords: Klebsiella pneumoniae; capsule polysaccharide; gut colonization; type VI secretion system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae.Front Cell Infect Microbiol. 2018 Jan 22;8:4. doi: 10.3389/fcimb.2018.00004. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29404282 Free PMC article. Review.
-
Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients.Clin Infect Dis. 2017 Jul 15;65(2):208-215. doi: 10.1093/cid/cix270. Clin Infect Dis. 2017. PMID: 28369261 Free PMC article.
-
Gastrointestinal carriage of Klebsiella pneumoniae in a general adult population: a cross-sectional study of risk factors and bacterial genomic diversity.Gut Microbes. 2021 Jan-Dec;13(1):1939599. doi: 10.1080/19490976.2021.1939599. Gut Microbes. 2021. PMID: 34182896 Free PMC article.
-
Deciphering the gastrointestinal carriage of Klebsiella pneumoniae.Infect Immun. 2024 Sep 10;92(9):e0048223. doi: 10.1128/iai.00482-23. Epub 2024 Apr 10. Infect Immun. 2024. PMID: 38597634 Free PMC article. Review.
-
Animal Model To Study Klebsiella pneumoniae Gastrointestinal Colonization and Host-to-Host Transmission.Infect Immun. 2020 Oct 19;88(11):e00071-20. doi: 10.1128/IAI.00071-20. Print 2020 Oct 19. Infect Immun. 2020. PMID: 32839189 Free PMC article.
Cited by
-
The Impact of Complementary Feeding on Fecal Microbiota in Exclusively Breast-Fed Infants with Cystic Fibrosis (A Descriptive Study).Nutrients. 2024 Nov 27;16(23):4071. doi: 10.3390/nu16234071. Nutrients. 2024. PMID: 39683464 Free PMC article.
-
Klebsiella pneumoniae evolution in the gut leads to spontaneous capsule loss and decreased virulence potential.mBio. 2025 May 14;16(5):e0236224. doi: 10.1128/mbio.02362-24. Epub 2025 Mar 31. mBio. 2025. PMID: 40162782 Free PMC article.
-
Klebsiella pneumoniae employs a type VI secretion system to overcome microbiota-mediated colonization resistance.Nat Commun. 2025 Jan 22;16(1):940. doi: 10.1038/s41467-025-56309-8. Nat Commun. 2025. PMID: 39843522 Free PMC article.
-
Leveraging collateral sensitivity to counteract the evolution of bacteriophage resistance in bacteria.mLife. 2025 Mar 18;4(2):143-154. doi: 10.1002/mlf2.70003. eCollection 2025 Apr. mLife. 2025. PMID: 40313983 Free PMC article.
-
Pseudomonas aeruginosa population dynamics in a vancomycin-induced murine model of gastrointestinal carriage.mBio. 2025 May 14;16(5):e0313624. doi: 10.1128/mbio.03136-24. Epub 2025 Apr 10. mBio. 2025. PMID: 40207916 Free PMC article.
References
-
- Gorrie CL, Mirceta M, Wick RR, Edwards DJ, Thomson NR, Strugnell RA, Pratt NF, Garlick JS, Watson KM, Pilcher DV, McGloughlin SA, Spelman DW, Jenney AWJ, Holt KE. 2017. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis 65:208–215. doi:10.1093/cid/cix270. - DOI - PMC - PubMed
-
- Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. The LancetInfectious Diseases 18:37–46. doi:10.1016/S1473-3099(17)30489-9. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
