Mycobacterium tuberculosis Requires the Outer Membrane Lipid Phthiocerol Dimycocerosate for Starvation-Induced Antibiotic Tolerance
- PMID: 36598240
- PMCID: PMC9948706
- DOI: 10.1128/msystems.00699-22
Mycobacterium tuberculosis Requires the Outer Membrane Lipid Phthiocerol Dimycocerosate for Starvation-Induced Antibiotic Tolerance
Abstract
Tolerance of Mycobacterium tuberculosis to antibiotics contributes to the long duration of tuberculosis (TB) treatment and the emergence of drug-resistant strains. M. tuberculosis drug tolerance is induced by nutrient restriction, but the genetic determinants that promote antibiotic tolerance triggered by nutrient limitation have not been comprehensively identified. Here, we show that M. tuberculosis requires production of the outer membrane lipid phthiocerol dimycocerosate (PDIM) to tolerate antibiotics under nutrient-limited conditions. We developed an arrayed transposon (Tn) mutant library in M. tuberculosis Erdman and used orthogonal pooling and transposon sequencing (Tn-seq) to map the locations of individual mutants in the library. We screened a subset of the library (~1,000 mutants) by Tn-seq and identified 32 and 102 Tn mutants with altered tolerance to antibiotics under stationary-phase and phosphate-starved conditions, respectively. Two mutants recovered from the arrayed library, ppgK::Tn and clpS::Tn, showed increased susceptibility to two different drug combinations under both nutrient-limited conditions, but their phenotypes were not complemented by the Tn-disrupted gene. Whole-genome sequencing revealed single nucleotide polymorphisms in both the ppgK::Tn and clpS::Tn mutants that prevented PDIM production. Complementation of the clpS::Tn ppsD Q291* mutant with ppsD restored PDIM production and antibiotic tolerance, demonstrating that loss of PDIM sensitized M. tuberculosis to antibiotics. Our data suggest that drugs targeting production of PDIM, a critical M. tuberculosis virulence determinant, have the potential to enhance the efficacy of existing antibiotics, thereby shortening TB treatment and limiting development of drug resistance. IMPORTANCE Mycobacterium tuberculosis causes 10 million cases of active TB disease and over 1 million deaths worldwide each year. TB treatment is complex, requiring at least 6 months of therapy with a combination of antibiotics. One factor that contributes to the length of TB treatment is M. tuberculosis phenotypic antibiotic tolerance, which allows the bacteria to survive prolonged drug exposure even in the absence of genetic mutations causing drug resistance. Here, we report a genetic screen to identify M. tuberculosis genes that promote drug tolerance during nutrient starvation. Our study revealed the outer membrane lipid phthiocerol dimycocerosate (PDIM) as a key determinant of M. tuberculosis antibiotic tolerance triggered by nutrient starvation. Our study implicates PDIM synthesis as a potential target for development of new TB drugs that would sensitize M. tuberculosis to existing antibiotics to shorten TB treatment.
Keywords: Mycobacterium tuberculosis; PDIM; Tn-seq; antibiotic resistance; drug tolerance; membrane permeability; nutrient limitation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
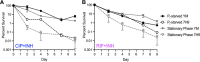
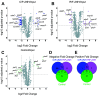
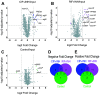
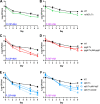

References
-
- WHO. 2022. WHO consolidated guidelines on tuberculosis. Module 4: treatment - drug-susceptible tuberculosis treatment. WHO, Geneva, Switzerland. - PubMed
-
- Sebastian J, Swaminath S, Nair RR, Jakkala K, Pradhan A, Ajitkumar P. 2017. De novo emergence of genetically resistant mutants of Mycobacterium tuberculosis from the persistence phase cells formed against antituberculosis drugs in vitro. Antimicrob Agents Chemother 61:e01343-16. doi:10.1128/AAC.01343-16. - DOI - PMC - PubMed
-
- WHO. 2021. Global tuberculosis report 2021. WHO, Geneva, Switzerland.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

