Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2
- PMID: 36598939
- PMCID: PMC9926234
- DOI: 10.1073/pnas.2212931120
Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2
Abstract
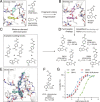
The nonstructural protein 3 (NSP3) of the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) contains a conserved macrodomain enzyme (Mac1) that is critical for pathogenesis and lethality. While small-molecule inhibitors of Mac1 have great therapeutic potential, at the outset of the COVID-19 pandemic, there were no well-validated inhibitors for this protein nor, indeed, the macrodomain enzyme family, making this target a pharmacological orphan. Here, we report the structure-based discovery and development of several different chemical scaffolds exhibiting low- to sub-micromolar affinity for Mac1 through iterations of computer-aided design, structural characterization by ultra-high-resolution protein crystallography, and binding evaluation. Potent scaffolds were designed with in silico fragment linkage and by ultra-large library docking of over 450 million molecules. Both techniques leverage the computational exploration of tangible chemical space and are applicable to other pharmacological orphans. Overall, 160 ligands in 119 different scaffolds were discovered, and 153 Mac1-ligand complex crystal structures were determined, typically to 1 Å resolution or better. Our analyses discovered selective and cell-permeable molecules, unexpected ligand-mediated conformational changes within the active site, and key inhibitor motifs that will template future drug development against Mac1.
Keywords: coronavirus; fragment-based drug discovery; macrodomain; virtual screening.
Conflict of interest statement
The authors have organizational affiliations to disclose, A.A. is a co-founder of Tango Therapeutics, Azkarra Therapeutics, and Ovibio Corporation; a consultant for SPARC, Bluestar, ProLynx, Earli, Cura, GenVivo, and GSK; a member of the SAB of Genentech, GLAdiator, Circle, and Cambridge Science Corporation; receives grant/research support from SPARC and AstraZeneca; and holds patents on the use of PARP inhibitors held jointly with AstraZeneca, which he has benefited financially (and may do so in the future). J.E.G. is consultant for Protego BioPharma and Contour Therapeutics as well as founder of Kaizen Therapeutics. B.K.S. is co-founder of BlueDolphin, LLC, a molecular docking contract research organization, Epiodyne, and Deep Apple Therapeutics, Inc., both drug discovery companies, has recently consulted for Umbra, Abbvie, and Dice Therapeutics, and is on the SAB of Schrodinger. J.J.I. co-founded Deep Apple Therapeutics, Inc., and BlueDolphin, LLC. J.S.F. is a consultant for, has equity in, and receives research support from Relay Therapeutics. I.A. is a consultant for Dark Blue Therapeutics., Yes, the authors have stock ownership to disclose, A.A. is a co-founder of Tango Therapeutics, Azkarra Therapeutics, and Ovibio Corporation; a consultant for SPARC, Bluestar, ProLynx, Earli, Cura, GenVivo, and GSK; a member of the SAB of Genentech, GLAdiator, Circle, and Cambridge Science Corporation; receives grant/research support from SPARC and AstraZeneca; and holds patents on the use of PARP inhibitors held jointly with AstraZeneca, which he has benefited financially (and may do so in the future). J.E.G. is consultant for Protego BioPharma and Contour Therapeutics as well as founder of Kaizen Therapeutics. B.K.S. is co-founder of BlueDolphin, LLC, a molecular docking contract research organization, Epiodyne, and Deep Apple Therapeutics, Inc., both drug discovery companies, has recently consulted for Umbra, Abbvie, and Dice Therapeutics, and is on the SAB of Schrodinger. J.J.I. co-founded Deep Apple Therapeutics, Inc., and BlueDolphin, LLC. J.S.F. is a consultant for, has equity in, and receives research support from Relay Therapeutics. I.A. is a consultant for Dark Blue Therapeutics.
Figures







Update of
-
Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 Macrodomain of SARS-CoV-2.bioRxiv [Preprint]. 2022 Jul 28:2022.06.27.497816. doi: 10.1101/2022.06.27.497816. bioRxiv. 2022. Update in: Proc Natl Acad Sci U S A. 2023 Jan 10;120(2):e2212931120. doi: 10.1073/pnas.2212931120. PMID: 35794891 Free PMC article. Updated. Preprint.
References
-
- Fu W., et al. , The search for inhibitors of macrodomains for targeting the readers and erasers of mono-ADP-ribosylation. Drug. Discov. Today 26, 2547–2558 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM141299/GM/NIGMS NIH HHS/United States
- P30 GM133893/GM/NIGMS NIH HHS/United States
- R01 GM071896/GM/NIGMS NIH HHS/United States
- T32 GM145460/GM/NIGMS NIH HHS/United States
- P30 GM124169/GM/NIGMS NIH HHS/United States
- R01 GM133836/GM/NIGMS NIH HHS/United States
- DH_/Department of Health/United Kingdom
- P30 GM133894/GM/NIGMS NIH HHS/United States
- 210634/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- BB/R007195/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R35 GM122481/GM/NIGMS NIH HHS/United States
- C35050/A22284/CRUK_/Cancer Research UK/United Kingdom
- 22284/CRUK_/Cancer Research UK/United Kingdom
- U19 AI171110/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

