Spleen tyrosine kinase inhibition restores myeloid homeostasis in COVID-19
- PMID: 36598976
- PMCID: PMC9812373
- DOI: 10.1126/sciadv.ade8272
Spleen tyrosine kinase inhibition restores myeloid homeostasis in COVID-19
Abstract
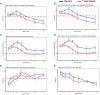
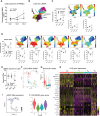
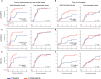
Spleen tyrosine kinase (SYK) is a previously unidentified therapeutic target that inhibits neutrophil and macrophage activation in coronavirus disease 2019 (COVID-19). Fostamatinib, a SYK inhibitor, was studied in a phase 2 placebo-controlled randomized clinical trial and was associated with improvements in many secondary end points related to efficacy. Here, we used a multiomic approach to evaluate cellular and soluble immune mediator responses of patients enrolled in this trial. We demonstrated that SYK inhibition was associated with reduced neutrophil activation, increased circulation of mature neutrophils (CD10+CD33-), and decreased circulation of low-density granulocytes and polymorphonuclear myeloid-derived suppressor cells (HLA-DR-CD33+CD11b-). SYK inhibition was also associated with normalization of transcriptional activity in circulating monocytes relative to healthy controls, an increase in frequency of circulating nonclassical and HLA-DRhi classical monocyte populations, and restoration of interferon responses. Together, these data suggest that SYK inhibition may mitigate proinflammatory myeloid cellular and soluble mediator responses thought to contribute to immunopathogenesis of severe COVID-19.
Figures



References
-
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

