An antisteatosis response regulated by oleic acid through lipid droplet-mediated ERAD enhancement
- PMID: 36598980
- PMCID: PMC9812393
- DOI: 10.1126/sciadv.adc8917
An antisteatosis response regulated by oleic acid through lipid droplet-mediated ERAD enhancement
Abstract
Although excessive lipid accumulation is a hallmark of obesity-related pathologies, some lipids are beneficial. Oleic acid (OA), the most abundant monounsaturated fatty acid (FA), promotes health and longevity. Here, we show that OA benefits Caenorhabditis elegans by activating the endoplasmic reticulum (ER)-resident transcription factor SKN-1A (Nrf1/NFE2L1) in a lipid homeostasis response. SKN-1A/Nrf1 is cleared from the ER by the ER-associated degradation (ERAD) machinery and stabilized when proteasome activity is low and canonically maintains proteasome homeostasis. Unexpectedly, OA increases nuclear SKN-1A levels independently of proteasome activity, through lipid droplet-dependent enhancement of ERAD. In turn, SKN-1A reduces steatosis by reshaping the lipid metabolism transcriptome and mediates longevity from OA provided through endogenous accumulation, reduced H3K4 trimethylation, or dietary supplementation. Our findings reveal an unexpected mechanism of FA signal transduction, as well as a lipid homeostasis pathway that provides strategies for opposing steatosis and aging, and may mediate some benefits of the OA-rich Mediterranean diet.
Figures


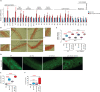


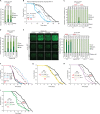
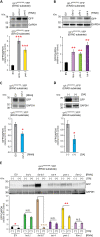

References
-
- H. Shimano, R. Sato, SREBP-regulated lipid metabolism: Convergent physiology-divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730 (2017). - PubMed
-
- H. Yki-Järvinen, P. K. Luukkonen, L. Hodson, J. B. Moore, Dietary carbohydrates and fats in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 18, 770–786 (2021). - PubMed
-
- D. D. Wang, F. B. Hu, Dietary fat and risk of cardiovascular disease: Recent controversies and advances. Annu. Rev. Nutr. 37, 423–446 (2017). - PubMed
-
- J. G. Wallis, J. L. Watts, J. Browse, Polyunsaturated fatty acid synthesis: What will they think of next? Trends Biochem. Sci. 27, 467–473 (2002). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

