In vitro cellular reprogramming to model gonad development and its disorders
- PMID: 36598988
- PMCID: PMC9812383
- DOI: 10.1126/sciadv.abn9793
In vitro cellular reprogramming to model gonad development and its disorders
Abstract
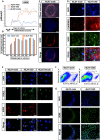
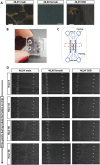
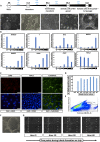
During embryonic development, mutually antagonistic signaling cascades determine gonadal fate toward a testicular or ovarian identity. Errors in this process result in disorders of sex development (DSDs), characterized by discordance between chromosomal, gonadal, and anatomical sex. The absence of an appropriate, accessible in vitro system is a major obstacle in understanding mechanisms of sex-determination/DSDs. Here, we describe protocols for differentiation of mouse and human pluripotent cells toward gonadal progenitors. Transcriptomic analysis reveals that the in vitro-derived murine gonadal cells are equivalent to embryonic day 11.5 in vivo progenitors. Using similar conditions, Sertoli-like cells derived from 46,XY human induced pluripotent stem cells (hiPSCs) exhibit sustained expression of testis-specific genes, secrete anti-Müllerian hormone, migrate, and form tubular structures. Cells derived from 46,XY DSD female hiPSCs, carrying an NR5A1 variant, show aberrant gene expression and absence of tubule formation. CRISPR-Cas9-mediated variant correction rescued the phenotype. This is a robust tool to understand mechanisms of sex determination and model DSDs.
Figures




References
-
- Vidal V., Schedl A., Requirement of WT1 for gonad and adrenal development: Insights from transgenic animals. Endocr. Res. 26, 1075–1082 (2000). - PubMed
-
- Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D., Jaenisch R., WT-1 is required for early kidney development. Cell 74, 679–691 (1993). - PubMed
-
- Luo X., Ikeda Y., Lala D. S., Baity L. A., Meade J. C., Parker K. L., A cell-specific nuclear receptor plays essential roles in adrenal and gonadal development. Endocr. Res. 21, 517–524 (1995). - PubMed
-
- Luo X., Ikeda Y., Parker K. L., A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77, 481–490 (1994). - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

