Commensal microbiome promotes hair follicle regeneration by inducing keratinocyte HIF-1α signaling and glutamine metabolism
- PMID: 36598999
- PMCID: PMC9812389
- DOI: 10.1126/sciadv.abo7555
Commensal microbiome promotes hair follicle regeneration by inducing keratinocyte HIF-1α signaling and glutamine metabolism
Abstract
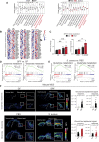
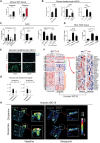
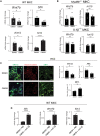
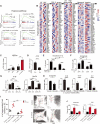
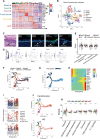
Tissue injury induces metabolic changes in stem cells, which likely modulate regeneration. Using a model of organ regeneration called wound-induced hair follicle neogenesis (WIHN), we identified skin-resident bacteria as key modulators of keratinocyte metabolism, demonstrating a positive correlation between bacterial load, glutamine metabolism, and regeneration. Specifically, through comprehensive multiomic analysis and single-cell RNA sequencing in murine skin, we show that bacterially induced hypoxia drives increased glutamine metabolism in keratinocytes with attendant enhancement of skin and hair follicle regeneration. In human skin wounds, topical broad-spectrum antibiotics inhibit glutamine production and are partially responsible for reduced healing. These findings reveal a conserved and coherent physiologic context in which bacterially induced metabolic changes improve the tolerance of stem cells to damage and enhance regenerative capacity. This unexpected proregenerative modulation of metabolism by the skin microbiome in both mice and humans suggests important methods for enhancing regeneration after injury.
Figures


References
-
- G. Wang, E. Sweren, H. Liu, E. Wier, M. P. Alphonse, R. Chen, N. Islam, A. Li, Y. Xue, J. Chen, Y. Chen, S. Lee, Y. Wang, S. Wang, N. K. Archer, W. Andrews, M. A. Kane, E. Dare, S. K. Reddy, Z. Hu, E. A. Grice, L. S. Miller, L. A. Garza, Bacteria induce skin regeneration via IL-1β signaling. Cell Host Microbe 29, 777–791.e6 (2021). - PMC - PubMed
-
- H. Abo, B. Chassaing, A. Harusato, M. Quiros, J. C. Brazil, V. L. Ngo, E. Viennois, D. Merlin, A. T. Gewirtz, A. Nusrat, T. L. Denning, Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration. Nat. Commun. 11, 513 (2020). - PMC - PubMed
-
- Y. Liu, Y. Wang, Y. Ni, C. K. Y. Cheung, K. S. L. Lam, Y. Wang, Z. Xia, D. Ye, J. Guo, M. A. Tse, G. Panagiotou, A. Xu, Gut microbiome fermentation determines the efficacy of exercise for diabetes prevention. Cell Metab. 31, 77–91.e5 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

