Immunoediting instructs tumor metabolic reprogramming to support immune evasion
- PMID: 36599297
- PMCID: PMC10375941
- DOI: 10.1016/j.cmet.2022.12.003
Immunoediting instructs tumor metabolic reprogramming to support immune evasion
Abstract
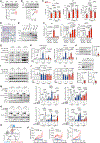
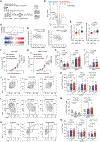
Immunoediting sculpts immunogenicity and thwarts host anti-tumor responses in tumor cells during tumorigenesis; however, it remains unknown whether metabolic programming of tumor cells can be guided by immunosurveillance. Here, we report that T cell-mediated immunosurveillance in early-stage tumorigenesis instructs c-Myc upregulation and metabolic reprogramming in tumor cells. This previously unexplored tumor-immune interaction is controlled by non-canonical interferon gamma (IFNγ)-STAT3 signaling and supports tumor immune evasion. Our findings uncover that immunoediting instructs deregulated bioenergetic programs in tumor cells to empower them to disarm the T cell-mediated immunosurveillance by imposing metabolic tug-of-war between tumor and infiltrating T cells and forming the suppressive tumor microenvironment.
Keywords: IFNγ; Myc; STAT3; immunoediting; immunosurveillance; tumor immunology.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests P.-C.H. is a member of the scientific advisory board for Elixiron Immunotherapeutics and received research grants from Elixiron Immunotherapeutics. P.-C.H. is also a founder of Pilatus Biosciences.
Figures




Comment in
-
Early immune pressure makes tumors metabolically stronger.Cell Metab. 2023 Jan 3;35(1):3-5. doi: 10.1016/j.cmet.2022.12.009. Cell Metab. 2023. PMID: 36599301
-
T cells feed into cancer immune escape.Nat Rev Cancer. 2023 Mar;23(3):113. doi: 10.1038/s41568-023-00550-0. Nat Rev Cancer. 2023. PMID: 36732446 No abstract available.
References
-
- Fendt SM, Frezza C, and Erez A (2020). Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov. 10, 1797–1807. 10.1158/2159-8290.CD-20-0844. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

