Neonatal imprinting of alveolar macrophages via neutrophil-derived 12-HETE
- PMID: 36599368
- PMCID: PMC9945843
- DOI: 10.1038/s41586-022-05660-7
Neonatal imprinting of alveolar macrophages via neutrophil-derived 12-HETE
Abstract
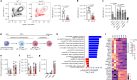
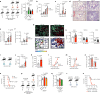
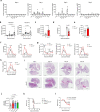
Resident-tissue macrophages (RTMs) arise from embryonic precursors1,2, yet the developmental signals that shape their longevity remain largely unknown. Here we demonstrate in mice genetically deficient in 12-lipoxygenase and 15-lipoxygenase (Alox15-/- mice) that neonatal neutrophil-derived 12-HETE is required for self-renewal and maintenance of alveolar macrophages (AMs) during lung development. Although the seeding and differentiation of AM progenitors remained intact, the absence of 12-HETE led to a significant reduction in AMs in adult lungs and enhanced senescence owing to increased prostaglandin E2 production. A compromised AM compartment resulted in increased susceptibility to acute lung injury induced by lipopolysaccharide and to pulmonary infections with influenza A virus or SARS-CoV-2. Our results highlight the complexity of prenatal RTM programming and reveal their dependency on in trans eicosanoid production by neutrophils for lifelong self-renewal.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

