An essential Noc3p dimerization cycle mediates ORC double-hexamer formation in replication licensing
- PMID: 36599624
- PMCID: PMC9813392
- DOI: 10.26508/lsa.202201594
An essential Noc3p dimerization cycle mediates ORC double-hexamer formation in replication licensing
Abstract
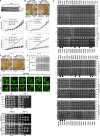
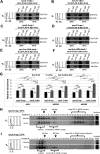
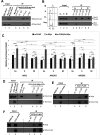
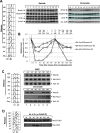
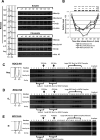
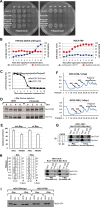
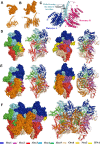
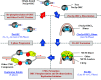
Replication licensing, a prerequisite of DNA replication, helps to ensure once-per-cell-cycle genome duplication. Some DNA replication-initiation proteins are sequentially loaded onto replication origins to form pre-replicative complexes (pre-RCs). ORC and Noc3p bind replication origins throughout the cell cycle, providing a platform for pre-RC assembly. We previously reported that cell cycle-dependent ORC dimerization is essential for the chromatin loading of the symmetric MCM double-hexamers. Here, we used Saccharomyces cerevisiae separation-of-function NOC3 mutants to confirm the separable roles of Noc3p in DNA replication and ribosome biogenesis. We also show that an essential and cell cycle-dependent Noc3p dimerization cycle regulates the ORC dimerization cycle. Noc3p dimerizes at the M-to-G1 transition and de-dimerizes in S-phase. The Noc3p dimerization cycle coupled with the ORC dimerization cycle enables replication licensing, protects nascent sister replication origins after replication initiation, and prevents re-replication. This study has revealed a new mechanism of replication licensing and elucidated the molecular mechanism of Noc3p as a mediator of ORC dimerization in pre-RC formation.
© 2023 Amin et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Amin A, Cheung MH, Liang C (2019) DNA replication-initiation proteins in eukaryotic cells. Biomed J Sci Tech Res 23: 17042–17049. 10.26717/BJSTR.2019.23.003830 - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
