An ISO-certified genomics workflow for identification and surveillance of antimicrobial resistance
- PMID: 36599823
- PMCID: PMC9813266
- DOI: 10.1038/s41467-022-35713-4
An ISO-certified genomics workflow for identification and surveillance of antimicrobial resistance
Abstract
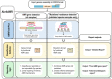
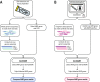
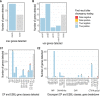
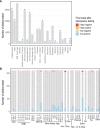
Realising the promise of genomics to revolutionise identification and surveillance of antimicrobial resistance (AMR) has been a long-standing challenge in clinical and public health microbiology. Here, we report the creation and validation of abritAMR, an ISO-certified bioinformatics platform for genomics-based bacterial AMR gene detection. The abritAMR platform utilises NCBI's AMRFinderPlus, as well as additional features that classify AMR determinants into antibiotic classes and provide customised reports. We validate abritAMR by comparing with PCR or reference genomes, representing 1500 different bacteria and 415 resistance alleles. In these analyses, abritAMR displays 99.9% accuracy, 97.9% sensitivity and 100% specificity. We also compared genomic predictions of phenotype for 864 Salmonella spp. against agar dilution results, showing 98.9% accuracy. The implementation of abritAMR in our institution has resulted in streamlined bioinformatics and reporting pathways, and has been readily updated and re-verified. The abritAMR tool and validation datasets are publicly available to assist laboratories everywhere harness the power of AMR genomics in professional practice.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- O’Neill J. Review on antimicrobial resistance: Tackling a crisis for the health and wealth of nations. London, UK: UK Government (2014).
-
- Centres for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S. Department of Health & Human Services (2019).
-
- World Health Organization. Global action plan on antimicrobial resistance. Geneva: WHO (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

