Transmembrane protein 135 regulates lipid homeostasis through its role in peroxisomal DHA metabolism
- PMID: 36599953
- PMCID: PMC9813353
- DOI: 10.1038/s42003-022-04404-7
Transmembrane protein 135 regulates lipid homeostasis through its role in peroxisomal DHA metabolism
Abstract
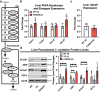
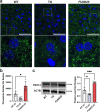
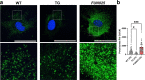
Transmembrane protein 135 (TMEM135) is thought to participate in the cellular response to increased intracellular lipids yet no defined molecular function for TMEM135 in lipid metabolism has been identified. In this study, we performed a lipid analysis of tissues from Tmem135 mutant mice and found striking reductions of docosahexaenoic acid (DHA) across all Tmem135 mutant tissues, indicating a role of TMEM135 in the production of DHA. Since all enzymes required for DHA synthesis remain intact in Tmem135 mutant mice, we hypothesized that TMEM135 is involved in the export of DHA from peroxisomes. The Tmem135 mutation likely leads to the retention of DHA in peroxisomes, causing DHA to be degraded within peroxisomes by their beta-oxidation machinery. This may lead to generation or alteration of ligands required for the activation of peroxisome proliferator-activated receptor a (PPARa) signaling, which in turn could result in increased peroxisomal number and beta-oxidation enzymes observed in Tmem135 mutant mice. We confirmed this effect of PPARa signaling by detecting decreased peroxisomes and their proteins upon genetic ablation of Ppara in Tmem135 mutant mice. Using Tmem135 mutant mice, we also validated the protective effect of increased peroxisomes and peroxisomal beta-oxidation on the metabolic disease phenotypes of leptin mutant mice which has been observed in previous studies. Thus, we conclude that TMEM135 has a role in lipid homeostasis through its function in peroxisomes.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

