CDK8/19 inhibition plays an important role in pancreatic β-cell induction from human iPSCs
- PMID: 36600289
- PMCID: PMC9814340
- DOI: 10.1186/s13287-022-03220-4
CDK8/19 inhibition plays an important role in pancreatic β-cell induction from human iPSCs
Abstract
Background: Transplantation of differentiated cells from human-induced pluripotent stem cells (hiPSCs) holds great promise for clinical treatments. Eliminating the risk factor of malignant cell transformation is essential for ensuring the safety of such cells. This study was aimed at assessing and mitigating mutagenicity that may arise during the cell culture process in the protocol of pancreatic islet cell (iPIC) differentiation from hiPSCs.
Methods: We evaluated the mutagenicity of differentiation factors used for hiPSC-derived pancreatic islet-like cells (iPICs). We employed Ames mutagenicity assay, flow cytometry analysis, immunostaining, time-resolved fluorescence resonance energy transfer-based (TR-FRET) cell-free dose-response assays, single-cell RNA-sequencing and in vivo efficacy study.
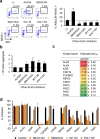
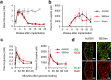
Results: We observed a mutagenic effect of activin receptor-like kinase 5 inhibitor II (ALK5iII). ALK5iII is a widely used β-cell inducer but no other tested ALK5 inhibitors induced β-cells. We obtained kinase inhibition profiles and found that only ALK5iII inhibited cyclin-dependent kinases 8 and 19 (CDK8/19) among all ALK5 inhibitors tested. Consistently, CDK8/19 inhibitors efficiently induced β-cells in the absence of ALK5iII. A combination treatment with non-mutagenic ALK5 inhibitor SB431542 and CDK8/19 inhibitor senexin B afforded generation of iPICs with in vitro cellular composition and in vivo efficacy comparable to those observed with ALK5iII.
Conclusion: Our findings suggest a new risk mitigation approach for cell therapy and advance our understanding of the β-cell differentiation mechanism.
Keywords: Activin receptor-like kinase 5 inhibitor II; CDK8/19 inhibitors; Human-induced pluripotent stem cells; Mutagenicity; Pancreatic islet cell.
© 2022. The Author(s).
Conflict of interest statement
Dr. Toyoda is a scientific advisor with advisory fee for Orizuru Therapeutics. Dr. Watanabe serves as a toxicological consultant for Orizuru but with no compensation. Drs. Sakuma, Yamazoe, Ueno, and Ito are employees of Orizuru. The remaining authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

