SP140 inhibits STAT1 signaling, induces IFN-γ in tumor-associated macrophages, and is a predictive biomarker of immunotherapy response
- PMID: 36600652
- PMCID: PMC9748993
- DOI: 10.1136/jitc-2022-005088
SP140 inhibits STAT1 signaling, induces IFN-γ in tumor-associated macrophages, and is a predictive biomarker of immunotherapy response
Abstract
Background: Understanding the role and potential therapeutic targeting of tumor-associated macrophages (TAMs) is crucial to developing new biomarkers and therapeutic strategies for cancer immunotherapies. The epigenetic reader SP140 has emerged as a master regulator of macrophage transcriptional programs; however, its role in the signaling of TAMs and response to immunotherapy has not been investigated.
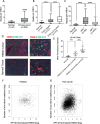
Methods: We evaluated the correlation between SP140 expression in head and neck squamous cell carcinoma (HNSCC) TAMs and clinical outcomes. We also used complementary bioinformatics and experimental approaches to study the association of SP140 expression with tumor mutation burden, patient survival, immunogenic signature of tumors, and signaling of TAMs. SP140 overexpression or knockdown was implemented to identify the role of SP140 in downstream signaling and production of inflammatory cytokine and chemokines. Chromatin immunoprecipitation and analysis of assay of transposase accessible chromatin sequencing data were used to demonstrate the direct binding of SP140 on the promoters of STAT1. Finally, correlation of SP140 with immune cell infiltrates and response to immune-checkpoint blockade in independent cohorts of HNSCC, metastatic melanoma, and melanoma was assessed.
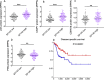
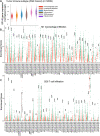
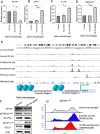
Results: We found that SP140 is highly expressed in TAMs across many cancer types, including HNSCCs. Interestingly, higher expression of SP140 in the tumors was associated with higher tumor mutation burden, improved survival, and a favorable response to immunotherapy. Tumors with high SP140 expression showed enrichment of inflammatory response and interferon-gamma (IFN-γ) pathways in both pan-cancer analysis and HNSCC-specific analysis. Mechanistically, SP140 negatively regulates transcription and phosphorylation of STAT1 and induces IFN-γ signaling. Activating SP140 in macrophages and TAMs induced the proinflammatory macrophage phenotype, increased the antitumor activity of macrophages, and increased the production of IFN-γ and antitumor cytokines and chemokines including interleukin-12 and CXCL10. SP140 expression provided higher sensitivity and specificity to predict antiprogrammed cell death protein 1 immunotherapy response compared with programmed death-ligand 1 in HNSCCs and lung cancer. In metastatic melanoma, higher levels of SP140 were associated with a durable response to immunotherapy, higher immune score estimates, high infiltrations of CD8+ T cells, and inflammatory TAMs.
Conclusions: Our findings suggest that SP140 could serve as both a therapeutic target and a biomarker to identify immunotherapy responders.
Keywords: biomarkers, tumor; head and neck neoplasms; immunotherapy; macrophages.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
