Macrophages and γδ T cells interplay during SARS-CoV-2 variants infection
- PMID: 36601113
- PMCID: PMC9806226
- DOI: 10.3389/fimmu.2022.1078741
Macrophages and γδ T cells interplay during SARS-CoV-2 variants infection
Abstract
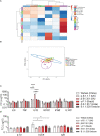
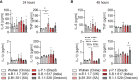
Introduction: The emergence of several SARS-CoV-2 variants during the COVID pandemic has revealed the impact of variant diversity on viral infectivity and host immune responses. While antibodies and CD8 T cells are essential to clear viral infection, the protective role of innate immunity including macrophages has been recognized. The aims of our study were to compare the infectivity of different SARS-CoV-2 variants in monocyte-derived macrophages (MDM) and to assess their activation profiles and the role of ACE2 (Angiotensin-converting enzyme 2), the main SARS-CoV-2 receptor. We also studied the ability of macrophages infected to affect other immune cells such as γδ2 T cells, another partner of innate immune response to viral infections.
Results: We showed that the SARS-CoV-2 variants α-B.1.1.7 (United Kingdom), β-B.1.351 (South Africa), γ-P.1 (Brazil), δ-B.1.617 (India) and B.1.1.529 (Omicron), infected MDM without replication, the γ-Brazil variant exhibiting increased infectivity for MDM. No clear polarization profile of SARS-CoV-2 variants-infected MDM was observed. The β-B.1.351 (South Africa) variant induced macrophage activation while B.1.1.529 (Omicron) was rather inhibitory. We observed that SARS-CoV-2 variants modulated ACE2 expression in MDM. In particular, the β-B.1.351 (South Africa) variant induced a higher expression of ACE2, related to MDM activation. Finally, all variants were able to activate γδ2 cells among which γ-P.1 (Brazil) and β-B.1.351 (South Africa) variants were the most efficient.
Conclusion: Our data show that SARS-CoV-2 variants can infect MDM and modulate their activation, which was correlated with the ACE2 expression. They also affect γδ2 T cell activation. The macrophage response to SARS-CoV-2 variants was stereotypical.
Keywords: ACE2; COVID-19; SARS-CoV-2 variants; macrophage; γδ T cells.
Copyright © 2022 Atmeh, Gay, Levasseur, La Scola, Olive, Mezouar, Gorvel and Mege.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

