Deep learning-based recognition and segmentation of intracranial aneurysms under small sample size
- PMID: 36601346
- PMCID: PMC9806214
- DOI: 10.3389/fphys.2022.1084202
Deep learning-based recognition and segmentation of intracranial aneurysms under small sample size
Abstract
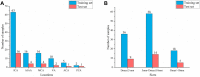
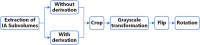
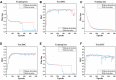
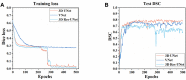
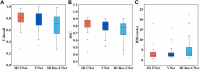
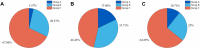
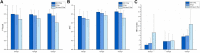
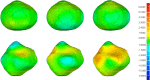
The manual identification and segmentation of intracranial aneurysms (IAs) involved in the 3D reconstruction procedure are labor-intensive and prone to human errors. To meet the demands for routine clinical management and large cohort studies of IAs, fast and accurate patient-specific IA reconstruction becomes a research Frontier. In this study, a deep-learning-based framework for IA identification and segmentation was developed, and the impacts of image pre-processing and convolutional neural network (CNN) architectures on the framework's performance were investigated. Three-dimensional (3D) segmentation-dedicated architectures, including 3D UNet, VNet, and 3D Res-UNet were evaluated. The dataset used in this study included 101 sets of anonymized cranial computed tomography angiography (CTA) images with 140 IA cases. After the labeling and image pre-processing, a training set and test set containing 112 and 28 IA lesions were used to train and evaluate the convolutional neural network mentioned above. The performances of three convolutional neural networks were compared in terms of training performance, segmentation performance, and segmentation efficiency using multiple quantitative metrics. All the convolutional neural networks showed a non-zero voxel-wise recall (V-Recall) at the case level. Among them, 3D UNet exhibited a better overall segmentation performance under the relatively small sample size. The automatic segmentation results based on 3D UNet reached an average V-Recall of 0.797 ± 0.140 (3.5% and 17.3% higher than that of VNet and 3D Res-UNet), as well as an average dice similarity coefficient (DSC) of 0.818 ± 0.100, which was 4.1%, and 11.7% higher than VNet and 3D Res-UNet. Moreover, the average Hausdorff distance (HD) of the 3D UNet was 3.323 ± 3.212 voxels, which was 8.3% and 17.3% lower than that of VNet and 3D Res-UNet. The three-dimensional deviation analysis results also showed that the segmentations of 3D UNet had the smallest deviation with a max distance of +1.4760/-2.3854 mm, an average distance of 0.3480 mm, a standard deviation (STD) of 0.5978 mm, a root mean square (RMS) of 0.7269 mm. In addition, the average segmentation time (AST) of the 3D UNet was 0.053s, equal to that of 3D Res-UNet and 8.62% shorter than VNet. The results from this study suggested that the proposed deep learning framework integrated with 3D UNet can provide fast and accurate IA identification and segmentation.
Keywords: SAH; automatic segmentation; convolutional neural network; deep learning; intracranial aneurysm.
Copyright © 2022 Zhu, Luo, Yang, Cai, Yeo, Yan and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
A benchmark study of convolutional neural networks in fully automatic segmentation of aortic root.Front Bioeng Biotechnol. 2023 Jun 15;11:1171868. doi: 10.3389/fbioe.2023.1171868. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37397959 Free PMC article.
-
Automatic segmentation and applicator reconstruction for CT-based brachytherapy of cervical cancer using 3D convolutional neural networks.J Appl Clin Med Phys. 2020 Oct;21(10):158-169. doi: 10.1002/acm2.13024. Epub 2020 Sep 29. J Appl Clin Med Phys. 2020. PMID: 32991783 Free PMC article.
-
An improved 3D-UNet-based brain hippocampus segmentation model based on MR images.BMC Med Imaging. 2024 Jul 5;24(1):166. doi: 10.1186/s12880-024-01346-w. BMC Med Imaging. 2024. PMID: 38970025 Free PMC article.
-
Attention-UNet architectures with pretrained backbones for multi-class cardiac MR image segmentation.Curr Probl Cardiol. 2024 Jan;49(1 Pt C):102129. doi: 10.1016/j.cpcardiol.2023.102129. Epub 2023 Oct 20. Curr Probl Cardiol. 2024. PMID: 37866419 Review.
-
Mini Review: Deep Learning for Atrial Segmentation From Late Gadolinium-Enhanced MRIs.Front Cardiovasc Med. 2020 May 27;7:86. doi: 10.3389/fcvm.2020.00086. eCollection 2020. Front Cardiovasc Med. 2020. PMID: 32528977 Free PMC article. Review.
Cited by
-
Integrating metagenomics and metabolomics to study the gut microbiome and host relationships in sports across different energy systems.Sci Rep. 2025 May 2;15(1):15356. doi: 10.1038/s41598-025-98973-2. Sci Rep. 2025. PMID: 40316630 Free PMC article.
-
Compare deep learning model and conventional logistic regression model for the identification of unstable saccular intracranial aneurysms in computed tomography angiography.Quant Imaging Med Surg. 2024 Apr 3;14(4):2993-3005. doi: 10.21037/qims-23-1732. Epub 2024 Mar 28. Quant Imaging Med Surg. 2024. PMID: 38617165 Free PMC article.
-
Global tendencies and frontier topics in hemodynamics research of intracranial aneurysms: a bibliometric analysis from 1999 to 2022.Front Physiol. 2023 Nov 21;14:1157787. doi: 10.3389/fphys.2023.1157787. eCollection 2023. Front Physiol. 2023. PMID: 38074335 Free PMC article.
-
Integrating PointNet-Based Model and Machine Learning Algorithms for Classification of Rupture Status of IAs.Bioengineering (Basel). 2024 Jun 28;11(7):660. doi: 10.3390/bioengineering11070660. Bioengineering (Basel). 2024. PMID: 39061742 Free PMC article.
-
Morphology and Texture-Guided Deep Neural Network for Intracranial Aneurysm Segmentation in 3D TOF-MRA.Neuroinformatics. 2024 Oct;22(4):731-744. doi: 10.1007/s12021-024-09683-5. Epub 2024 Sep 11. Neuroinformatics. 2024. PMID: 39259472
References
-
- Adams W. M., Laitt R. D., Jackson A. (2000). The role of mr angiography in the pretreatment assessment of intracranial aneurysms: A comparative study. AJNR. Am. J. Neuroradiol. 21, 1618–1628. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11039340. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

