Dynamic regulation and key roles of ribonucleic acid methylation
- PMID: 36601431
- PMCID: PMC9806184
- DOI: 10.3389/fncel.2022.1058083
Dynamic regulation and key roles of ribonucleic acid methylation
Abstract
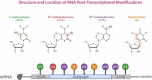
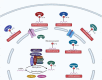
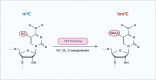
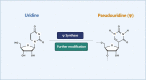
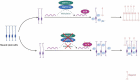
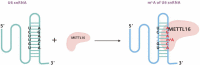
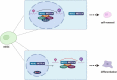
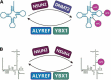
Ribonucleic acid (RNA) methylation is the most abundant modification in biological systems, accounting for 60% of all RNA modifications, and affects multiple aspects of RNA (including mRNAs, tRNAs, rRNAs, microRNAs, and long non-coding RNAs). Dysregulation of RNA methylation causes many developmental diseases through various mechanisms mediated by N 6-methyladenosine (m6A), 5-methylcytosine (m5C), N 1-methyladenosine (m1A), 5-hydroxymethylcytosine (hm5C), and pseudouridine (Ψ). The emerging tools of RNA methylation can be used as diagnostic, preventive, and therapeutic markers. Here, we review the accumulated discoveries to date regarding the biological function and dynamic regulation of RNA methylation/modification, as well as the most popularly used techniques applied for profiling RNA epitranscriptome, to provide new ideas for growth and development.
Keywords: 5-hydroxymethylcytosine; 5-methylcytosine; N1-methyladenosine; N6-methyladenosine; RNA methylation; pseudouridine.
Copyright © 2022 Zou, Liu, Tan, Chen, Dong, Bai, Wu and Zeng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Deciphering the epitranscriptome: A green perspective.J Integr Plant Biol. 2016 Oct;58(10):822-835. doi: 10.1111/jipb.12483. Epub 2016 Jun 20. J Integr Plant Biol. 2016. PMID: 27172004 Free PMC article. Review.
-
Role of Main RNA Methylation in Hepatocellular Carcinoma: N6-Methyladenosine, 5-Methylcytosine, and N1-Methyladenosine.Front Cell Dev Biol. 2021 Nov 30;9:767668. doi: 10.3389/fcell.2021.767668. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34917614 Free PMC article. Review.
-
Chemical Modifications to RNA: A New Layer of Gene Expression Regulation.ACS Chem Biol. 2017 Feb 17;12(2):316-325. doi: 10.1021/acschembio.6b00960. Epub 2017 Jan 12. ACS Chem Biol. 2017. PMID: 28051309 Review.
-
RNA modifications and their role in gene expression.Front Mol Biosci. 2025 Apr 25;12:1537861. doi: 10.3389/fmolb.2025.1537861. eCollection 2025. Front Mol Biosci. 2025. PMID: 40351534 Free PMC article. Review.
-
Decoding the Atlas of RNA Modifications from Epitranscriptome Sequencing Data.Methods Mol Biol. 2019;1870:107-124. doi: 10.1007/978-1-4939-8808-2_8. Methods Mol Biol. 2019. PMID: 30539550
Cited by
-
RNA Modification in Metabolism.MedComm (2020). 2025 Mar 10;6(3):e70135. doi: 10.1002/mco2.70135. eCollection 2025 Mar. MedComm (2020). 2025. PMID: 40066222 Free PMC article. Review.
-
Influence of RNA Methylation on Cancerous Cells: A Prospective Approach for Alteration of In Vivo Cellular Composition.Adv Exp Med Biol. 2025;1474:79-103. doi: 10.1007/5584_2024_820. Adv Exp Med Biol. 2025. PMID: 39259424 Review.
-
m 6A methylation in cellular senescence of age-associated diseases.Acta Biochim Biophys Sin (Shanghai). 2023 Jul 3;55(8):1168-1183. doi: 10.3724/abbs.2023107. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37394885 Free PMC article. Review.
References
-
- Ahmed S., Hossain Z., Uddin M., Taherzadeh G., Sharma A., Shatabda S., et al. (2020). Accurate prediction of Rna 5-hydroxymethylcytosine modification by utilizing novel position-specific gapped k-mer descriptors. Comput. Struct. Biotechnol. J. 18 3528–3538. 10.1016/j.csbj.2020.10.032 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

