Collective cell migration is spatiotemporally regulated during mammary epithelial bifurcation
- PMID: 36602106
- PMCID: PMC10112963
- DOI: 10.1242/jcs.259275
Collective cell migration is spatiotemporally regulated during mammary epithelial bifurcation
Abstract
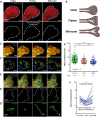
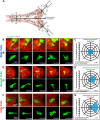
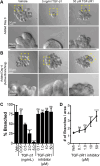
Branched epithelial networks are generated through an iterative process of elongation and bifurcation. We sought to understand bifurcation of the mammary epithelium. To visualize this process, we utilized three-dimensional (3D) organotypic culture and time-lapse confocal microscopy. We tracked cell migration during bifurcation and observed local reductions in cell speed at the nascent bifurcation cleft. This effect was proximity dependent, as individual cells approaching the cleft reduced speed, whereas cells exiting the cleft increased speed. As the cells slow down, they orient both migration and protrusions towards the nascent cleft, while cells in the adjacent branches orient towards the elongating tips. We next tested the hypothesis that TGF-β signaling controls mammary branching by regulating cell migration. We first validated that addition of TGF-β1 (TGFB1) protein increased cleft number, whereas inhibition of TGF-β signaling reduced cleft number. Then, consistent with our hypothesis, we observed that pharmacological inhibition of TGF-β1 signaling acutely decreased epithelial migration speed. Our data suggest a model for mammary epithelial bifurcation in which TGF-β signaling regulates cell migration to determine the local sites of bifurcation and the global pattern of the tubular network.
Keywords: Bifurcation; Branching morphogenesis; Collective cell migration; Epithelial development; Mammary gland; TGF-β.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests A.J.E. has unlicensed patents and patent applications related to the use of K14 as a prognostic biomarker, to the use of antibodies in cancer treatment, and to 3D culture protocols for tumor organoids. AJE’s spouse is an employee of Immunocore. All other authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

