Structural Analyses of a Dominant Cryptosporidium parvum Epitope Presented by H-2Kb Offer New Options To Combat Cryptosporidiosis
- PMID: 36602309
- PMCID: PMC9973275
- DOI: 10.1128/mbio.02666-22
Structural Analyses of a Dominant Cryptosporidium parvum Epitope Presented by H-2Kb Offer New Options To Combat Cryptosporidiosis
Abstract
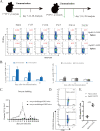
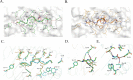
Cryptosporidium parvum has gained much attention as a major cause of diarrhea in the world, particularly in those with compromised immune systems. The data currently available on how the immune system recognizes C. parvum are growing rapidly, but we lack data on the interactions among host major histocompatibility complex (MHC) diversity and parasitic T-cell epitopes. To identify antigenic epitopes in a murine model, we performed systematic profiling of H-2Kb-restricted peptides by screening the dominant Cryptosporidium antigens. The results revealed that the glycoprotein-derived epitope Gp40/15-SVF9 induced an immunodominant response in C. parvum-recovered C57BL/6 mice, and injection of the cytotoxic-T-lymphocyte (CTL) peptide with the adjuvant activated peptide-specific CD8+ T cells. Notably, the SVF9 epitope was highly conserved across Cryptosporidium hominis, C. parvum, and many other Cryptosporidium species. SVF9 also formed stable peptide-MHC class I (MHC I) complexes with HLA-A*0201, suggesting cross-reactivity between H-2Kb and human MHC I specificities. Crystal structure analyses revealed that the interactions of peptide-MHC surface residues of H-2Kb and HLA-A*0201 are highly conserved. The hydrogen bonds of H-2Kb-SVF9 are similar to those of a dominant epitope presented by HLA-A*0201, which can be recognized by a public human T-cell receptor (TCR). Notably, we found double conformations in position 4 (P4), 5 (P5) of the SVF9 peptide, which showed high flexibility, and multiple peptide conformations generated more molecular surfaces that can potentially be recognized by TCRs. Our findings demonstrate that an immunodominant C. parvum epitope and its homologs from different Cryptosporidium species and subtypes can benefit vaccine development to combat cryptosporidiosis. IMPORTANCE Adaptive immune responses and T lymphocytes have been implicated as important mechanisms of parasite-induced protection. However, the role of CD8+ T lymphocytes in the resolution of C. parvum infection is largely unresolved. Our results revealed that the glycoprotein-derived epitope Gp40/15-SVF9 induced an immunodominant CD8+ T-cell response in C57BL/6 mice. Crystal structure analyses revealed that the interactions of the H-2Kb-SVF9 peptide are similar to those of a dominant epitope presented by HLA-A*0201, which can be recognized by human TCRs. In addition, we found double conformations of the SVF9 peptide, which showed high flexibility and multiple peptide conformations that can potentially be recognized by TCRs.
Keywords: Cryptosporidium parvum; MHC I; T-cell epitope; cell-mediated immunity.
Conflict of interest statement
We declare no conflict of interest.
Figures






Similar articles
-
Structural analyses of Cryptosporidium parvum epitopes reveal a novel scheme of decapeptide binding to H-2Kb.J Struct Biol. 2025 Mar;217(1):108168. doi: 10.1016/j.jsb.2025.108168. Epub 2025 Jan 12. J Struct Biol. 2025. PMID: 39809366
-
The signal sequence of lymphocytic choriomeningitis virus contains an immunodominant cytotoxic T cell epitope that is restricted by both H-2D(b) and H-2K(b) molecules.Virology. 1997 Jul 21;234(1):62-73. doi: 10.1006/viro.1997.8627. Virology. 1997. PMID: 9234947
-
Identification of the glycopeptide epitope recognized by a protective Cryptosporidium monoclonal antibody.Infect Immun. 2023 Oct 17;91(10):e0027523. doi: 10.1128/iai.00275-23. Epub 2023 Sep 19. Infect Immun. 2023. PMID: 37725059 Free PMC article.
-
[MHC tetramers: tracking specific immunity].Acta Med Croatica. 2003;57(4):255-9. Acta Med Croatica. 2003. PMID: 14639858 Review. Croatian.
-
Innate and cell-mediated immune responses to Cryptosporidium parvum.Adv Parasitol. 1998;40:87-119. doi: 10.1016/s0065-308x(08)60118-9. Adv Parasitol. 1998. PMID: 9554071 Review.
Cited by
-
Genetic crosses reveal genomic loci responsible for virulence in Cryptosporidium parvum infection.bioRxiv [Preprint]. 2025 May 21:2025.05.20.655157. doi: 10.1101/2025.05.20.655157. bioRxiv. 2025. PMID: 40475546 Free PMC article. Preprint.
-
Dendritic cell-mediated responses to secreted Cryptosporidium effectors promote parasite-specific CD8+ T cell responses.Mucosal Immunol. 2024 Jun;17(3):387-401. doi: 10.1016/j.mucimm.2024.03.003. Epub 2024 Mar 18. Mucosal Immunol. 2024. PMID: 38508522 Free PMC article.
-
Immunity to Cryptosporidium: insights into principles of enteric responses to infection.Nat Rev Immunol. 2024 Feb;24(2):142-155. doi: 10.1038/s41577-023-00932-3. Epub 2023 Sep 11. Nat Rev Immunol. 2024. PMID: 37697084 Free PMC article. Review.
References
-
- Checkley W, White AC, Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen X-M, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA, Jr, Priest JW, Roos DS, Striepen B, Thompson RCA, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis 15:85–94. doi:10.1016/S1473-3099(14)70772-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
