Convergent Community Assembly among Globally Separated Acidic Cave Biofilms
- PMID: 36602326
- PMCID: PMC9888236
- DOI: 10.1128/aem.01575-22
Convergent Community Assembly among Globally Separated Acidic Cave Biofilms
Abstract
Acidophilic bacteria and archaea inhabit extreme geochemical "islands" that can tell us when and how geographic barriers affect the biogeography of microorganisms. Here, we describe microbial communities from extremely acidic (pH 0 to 1) biofilms, known as snottites, from hydrogen sulfide-rich caves. Given the extreme acidity and subsurface location of these biofilms, and in light of earlier work showing strong geographic patterns among snottite Acidithiobacillus populations, we investigated their structure and diversity in order to understand how geography might impact community assembly. We used 16S rRNA gene cloning and fluorescence in situ hybridization (FISH) to investigate 26 snottite samples from four sulfidic caves in Italy and Mexico. All samples had very low biodiversity and were dominated by sulfur-oxidizing bacteria in the genus Acidithiobacillus. Ferroplasma and other archaea in the Thermoplasmatales ranged from 0 to 50% of total cells, and relatives of the bacterial genera Acidimicrobium and Ferrimicrobium were up to 15% of total cells. Rare phylotypes included Sulfobacillus spp. and members of the phyla "Candidatus Dependentiae" and "Candidatus Saccharibacteria" (formerly TM6 and TM7). Although the same genera of acidophiles occurred in snottites on separate continents, most members of those genera represent substantially divergent populations, with 16S rRNA genes that are only 95 to 98% similar. Our findings are consistent with a model of community assembly where sulfidic caves are stochastically colonized by microorganisms from local sources, which are strongly filtered through environmental selection for extreme acid tolerance, and these different colonization histories are maintained by dispersal restrictions within and among caves. IMPORTANCE Microorganisms that are adapted to extremely acidic conditions, known as extreme acidophiles, are catalysts for rock weathering, metal cycling, and mineral formation in naturally acidic environments. They are also important drivers of large-scale industrial processes such as biomining and contaminant remediation. Understanding the factors that govern their ecology and distribution can help us better predict and utilize their activities in natural and engineered systems. However, extremely acidic habitats are unusual in that they are almost always isolated within circumneutral landscapes. So where did their acid-adapted inhabitants come from, and how do new colonists arrive and become established? In this study, we took advantage of a unique natural experiment in Earth's subsurface to show how isolation may have played a role in the colonization history, community assembly, and diversity of highly acidic microbial biofilms.
Keywords: Acidithiobacillus; acidophiles; biofilms; biogeography; cave.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




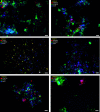

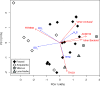
References
-
- Rowe OF, Sánchez-España J, Hallberg KB, Johnson DB. 2007. Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ Microbiol 9:1761–1771. 10.1111/j.1462-2920.2007.01294.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

