Heterozygous mutations in SOX2 may cause idiopathic hypogonadotropic hypogonadism via dominant-negative mechanisms
- PMID: 36602867
- PMCID: PMC9977424
- DOI: 10.1172/jci.insight.164324
Heterozygous mutations in SOX2 may cause idiopathic hypogonadotropic hypogonadism via dominant-negative mechanisms
Abstract
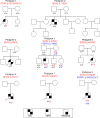
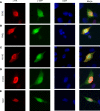
Pathogenic SRY-box transcription factor 2 (SOX2) variants typically cause severe ocular defects within a SOX2 disorder spectrum that includes hypogonadotropic hypogonadism. We examined exome-sequencing data from a large, well-phenotyped cohort of patients with idiopathic hypogonadotropic hypogonadism (IHH) for pathogenic SOX2 variants to investigate the underlying pathogenic SOX2 spectrum and its associated phenotypes. We identified 8 IHH individuals harboring heterozygous pathogenic SOX2 variants with variable ocular phenotypes. These variant proteins were tested in vitro to determine whether a causal relationship between IHH and SOX2 exists. We found that Sox2 was highly expressed in the hypothalamus of adult mice and colocalized with kisspeptin 1 (KISS1) expression in the anteroventral periventricular nucleus of adult female mice. In vitro, shRNA suppression of mouse SOX2 protein in Kiss-expressing cell lines increased the levels of human kisspeptin luciferase (hKiss-luc) transcription, while SOX2 overexpression repressed hKiss-luc transcription. Further, 4 of the identified SOX2 variants prevented this SOX2-mediated repression of hKiss-luc. Together, these data suggest that pathogenic SOX2 variants contribute to both anosmic and normosmic forms of IHH, attesting to hypothalamic defects in the SOX2 disorder spectrum. Our study describes potentially novel mechanisms contributing to SOX2-related disease and highlights the necessity of SOX2 screening in IHH genetic evaluation irrespective of associated ocular defects.
Keywords: Endocrinology; Neuroendocrine regulation; Neuroscience.
Figures




References
-
- Williamson KA, et al. SOX2 Disorder. In: Adam MP, et al., eds. GeneReviews. University of Washington; 2022. https://www.ncbi.nlm.nih.gov/books/NBK1300/ - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- P50 HD028138/HD/NICHD NIH HHS/United States
- R01 HD100580/HD/NICHD NIH HHS/United States
- K12 GM068524/GM/NIGMS NIH HHS/United States
- P42 ES010337/ES/NIEHS NIH HHS/United States
- R37 HD043341/HD/NICHD NIH HHS/United States
- R01 DK111038/DK/NIDDK NIH HHS/United States
- T32 AI007476/AI/NIAID NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- F32 HD090837/HD/NICHD NIH HHS/United States
- F32 HD108873/HD/NICHD NIH HHS/United States
- P50 HD012303/HD/NICHD NIH HHS/United States
- M01 RR000125/RR/NCRR NIH HHS/United States
- P50 HD104224/HD/NICHD NIH HHS/United States
- R01 HD096324/HD/NICHD NIH HHS/United States
- R01 HD072754/HD/NICHD NIH HHS/United States
- R01 HD082567/HD/NICHD NIH HHS/United States
- T32 HD007203/HD/NICHD NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- R01 DE031452/DE/NIDCR NIH HHS/United States
- R01 HD043341/HD/NICHD NIH HHS/United States
- K99 NS119291/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

