MYB insufficiency disrupts proteostasis in hematopoietic stem cells, leading to age-related neoplasia
- PMID: 36603185
- PMCID: PMC10646772
- DOI: 10.1182/blood.2022019138
MYB insufficiency disrupts proteostasis in hematopoietic stem cells, leading to age-related neoplasia
Abstract
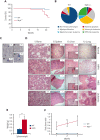
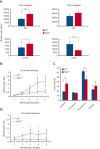
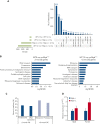
MYB plays a key role in gene regulation throughout the hematopoietic hierarchy and is critical for the maintenance of normal hematopoietic stem cells (HSC). Acquired genetic dysregulation of MYB is involved in the etiology of a number of leukemias, although inherited noncoding variants of the MYB gene are a susceptibility factor for many hematological conditions, including myeloproliferative neoplasms (MPN). The mechanisms that connect variations in MYB levels to disease predisposition, especially concerning age dependency in disease initiation, are completely unknown. Here, we describe a model of Myb insufficiency in mice that leads to MPN, myelodysplasia, and leukemia in later life, mirroring the age profile of equivalent human diseases. We show that this age dependency is intrinsic to HSC, involving a combination of an initial defective cellular state resulting from small effects on the expression of multiple genes and a progressive accumulation of further subtle changes. Similar to previous studies showing the importance of proteostasis in HSC maintenance, we observed altered proteasomal activity and elevated proliferation indicators, followed by elevated ribosome activity in young Myb-insufficient mice. We propose that these alterations combine to cause an imbalance in proteostasis, potentially creating a cellular milieu favoring disease initiation.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures


Comment in
-
MYB deficiency leads to myeloid neoplasia too.Blood. 2023 Apr 13;141(15):1786-1787. doi: 10.1182/blood.2022019533. Blood. 2023. PMID: 37052943 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

