Microbiota-dependent proteolysis of gluten subverts diet-mediated protection against type 1 diabetes
- PMID: 36603588
- PMCID: PMC9911364
- DOI: 10.1016/j.chom.2022.12.009
Microbiota-dependent proteolysis of gluten subverts diet-mediated protection against type 1 diabetes
Abstract
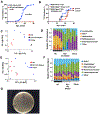
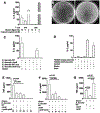
Diet and commensals can affect the development of autoimmune diseases like type 1 diabetes (T1D). However, whether dietary interventions are microbe-mediated was unclear. We found that a diet based on hydrolyzed casein (HC) as a protein source protects non-obese diabetic (NOD) mice in conventional and germ-free (GF) conditions via improvement in the physiology of insulin-producing cells to reduce autoimmune activation. The addition of gluten (a cereal protein complex associated with celiac disease) facilitates autoimmunity dependent on microbial proteolysis of gluten: T1D develops in GF animals monocolonized with Enterococcus faecalis harboring secreted gluten-digesting proteases but not in mice colonized with protease deficient bacteria. Gluten digestion by E. faecalis generates T cell-activating peptides and promotes innate immunity by enhancing macrophage reactivity to lipopolysaccharide (LPS). Gnotobiotic NOD Toll4-negative mice monocolonized with E. faecalis on an HC + gluten diet are resistant to T1D. These findings provide insights into strategies to develop dietary interventions to help protect humans against autoimmunity.
Keywords: celiac disease; diet and autoimmunity; insulin secretion regulation; microbial proteolysis of gluten; microbiota and autoimmunity; type 1 diabetes.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
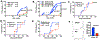



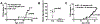
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

