Noncanonical DNA structures are drivers of genome evolution
- PMID: 36604282
- PMCID: PMC9877202
- DOI: 10.1016/j.tig.2022.11.005
Noncanonical DNA structures are drivers of genome evolution
Abstract
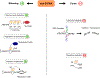
In addition to the canonical right-handed double helix, other DNA structures, termed 'non-B DNA', can form in the genomes across the tree of life. Non-B DNA regulates multiple cellular processes, including replication and transcription, yet its presence is associated with elevated mutagenicity and genome instability. These discordant cellular roles fuel the enormous potential of non-B DNA to drive genomic and phenotypic evolution. Here we discuss recent studies establishing non-B DNA structures as novel functional elements subject to natural selection, affecting evolution of transposable elements (TEs), and specifying centromeres. By highlighting the contributions of non-B DNA to repeated evolution and adaptation to changing environments, we conclude that evolutionary analyses should include a perspective of not only DNA sequence, but also its structure.
Keywords: G-quadruplexes; Z-DNA; mutations; natural selection; noncanonical DNA structure.
Copyright © 2022 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests No interests are declared.
Figures
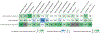
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

