3D Printing of Self-Assembling Nanofibrous Multidomain Peptide Hydrogels
- PMID: 36604310
- PMCID: PMC10023392
- DOI: 10.1002/adma.202210378
3D Printing of Self-Assembling Nanofibrous Multidomain Peptide Hydrogels
Abstract
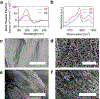
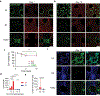
3D printing has become one of the primary fabrication strategies used in biomedical research. Recent efforts have focused on the 3D printing of hydrogels to create structures that better replicate the mechanical properties of biological tissues. These pose a unique challenge, as soft materials are difficult to pattern in three dimensions with high fidelity. Currently, a small number of biologically derived polymers that form hydrogels are frequently reused for 3D printing applications. Thus, there exists a need for novel hydrogels with desirable biological properties that can be used as 3D printable inks. In this work, the printability of multidomain peptides (MDPs), a class of self-assembling peptides that form a nanofibrous hydrogel at low concentrations, is established. MDPs with different charge functionalities are optimized as distinct inks and are used to create complex 3D structures, including multi-MDP prints. Additionally, printed MDP constructs are used to demonstrate charge-dependent differences in cellular behavior in vitro. This work presents the first time that self-assembling peptides have been used to print layered structures with overhangs and internal porosity. Overall, MDPs are a promising new class of 3D printable inks that are uniquely peptide-based and rely solely on supramolecular mechanisms for assembly.
Keywords: 3D printing; biomaterials; hydrogels; self-assembling peptides; supramolecular chemistry.
© 2023 Wiley-VCH GmbH.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures



Similar articles
-
Bioprinting synthetic self-assembling peptide hydrogels for biomedical applications.Biomed Mater. 2015 Dec 23;11(1):014103. doi: 10.1088/1748-6041/11/1/014103. Biomed Mater. 2015. PMID: 26694103 Review.
-
Composite Inks for Extrusion Printing of Biological and Biomedical Constructs.ACS Biomater Sci Eng. 2021 Sep 13;7(9):4009-4026. doi: 10.1021/acsbiomaterials.0c01158. Epub 2020 Nov 10. ACS Biomater Sci Eng. 2021. PMID: 34510905 Review.
-
3D printing of self-standing and vascular supportive multimaterial hydrogel structures for organ engineering.Biotechnol Bioeng. 2022 Jan;119(1):118-133. doi: 10.1002/bit.27954. Epub 2021 Oct 14. Biotechnol Bioeng. 2022. PMID: 34617587
-
Recent advances in high-strength and elastic hydrogels for 3D printing in biomedical applications.Acta Biomater. 2019 Sep 1;95:50-59. doi: 10.1016/j.actbio.2019.05.032. Epub 2019 May 22. Acta Biomater. 2019. PMID: 31125728 Free PMC article. Review.
-
3D Printing of Supramolecular Polymer Hydrogels with Hierarchical Structure.Small. 2021 Feb;17(5):e2005743. doi: 10.1002/smll.202005743. Epub 2021 Jan 15. Small. 2021. PMID: 33448102
Cited by
-
Nanofiber Scaffolds as Drug Delivery Systems Promoting Wound Healing.Pharmaceutics. 2023 Jun 26;15(7):1829. doi: 10.3390/pharmaceutics15071829. Pharmaceutics. 2023. PMID: 37514015 Free PMC article. Review.
-
Tunable Macroscopic Alignment of Self-Assembling Peptide Nanofibers.bioRxiv [Preprint]. 2024 Feb 4:2024.02.02.578651. doi: 10.1101/2024.02.02.578651. bioRxiv. 2024. Update in: ACS Nano. 2024 May 14;18(19):12477-12488. doi: 10.1021/acsnano.4c02030. PMID: 38352501 Free PMC article. Updated. Preprint.
-
High-Performance Sunlight-Induced Polymerized Hydrogels and Applications in 3D and 4D Printing.Small. 2025 Feb;21(5):e2411888. doi: 10.1002/smll.202411888. Epub 2024 Dec 18. Small. 2025. PMID: 39696970 Free PMC article.
-
Orthogonal Self-Assembly of Amphiphilic Peptide Hydrogels and Liposomes Results in Composite Materials with Tunable Release Profiles.Biomacromolecules. 2023 Nov 13;24(11):5018-5026. doi: 10.1021/acs.biomac.3c00664. Epub 2023 Sep 10. Biomacromolecules. 2023. PMID: 37690094 Free PMC article.
-
Recent Advances in the Development of Biomimetic Materials.Gels. 2023 Oct 20;9(10):833. doi: 10.3390/gels9100833. Gels. 2023. PMID: 37888406 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

