Human TH17 cells engage gasdermin E pores to release IL-1α on NLRP3 inflammasome activation
- PMID: 36604548
- PMCID: PMC9892007
- DOI: 10.1038/s41590-022-01386-w
Human TH17 cells engage gasdermin E pores to release IL-1α on NLRP3 inflammasome activation
Abstract
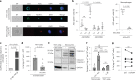
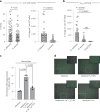
It has been shown that innate immune responses can adopt adaptive properties such as memory. Whether T cells utilize innate immune signaling pathways to diversify their repertoire of effector functions is unknown. Gasdermin E (GSDME) is a membrane pore-forming molecule that has been shown to execute pyroptotic cell death and thus to serve as a potential cancer checkpoint. In the present study, we show that human T cells express GSDME and, surprisingly, that this expression is associated with durable viability and repurposed for the release of the alarmin interleukin (IL)-1α. This property was restricted to a subset of human helper type 17 T cells with specificity for Candida albicans and regulated by a T cell-intrinsic NLRP3 inflammasome, and its engagement of a proteolytic cascade of successive caspase-8, caspase-3 and GSDME cleavage after T cell receptor stimulation and calcium-licensed calpain maturation of the pro-IL-1α form. Our results indicate that GSDME pore formation in T cells is a mechanism of unconventional cytokine release. This finding diversifies our understanding of the functional repertoire and mechanistic equipment of T cells and has implications for antifungal immunity.
© 2023. The Author(s).
Conflict of interest statement
A patent application has been filed by the authors. Otherwise, the authors declare no competing interests.
Figures






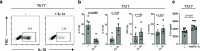









References
-
- Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. - PubMed
-
- Stockinger B, Omenetti S. The dichotomous nature of T helper 17 cells. Nat. Rev. Immunol. 2017;17:535–544. - PubMed
-
- Zielinski CE, et al. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–518. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

