OASL phase condensation induces amyloid-like fibrillation of RIPK3 to promote virus-induced necroptosis
- PMID: 36604592
- PMCID: PMC9859756
- DOI: 10.1038/s41556-022-01039-y
OASL phase condensation induces amyloid-like fibrillation of RIPK3 to promote virus-induced necroptosis
Abstract
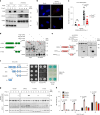
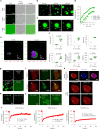
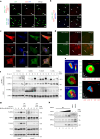
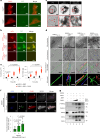
RIPK3-ZBP1-MLKL-mediated necroptosis is a proinflammatory cell death process that is crucial for antiviral host defence. RIPK3 self-oligomerization and autophosphorylation are prerequisites for executing necroptosis, yet the underlying mechanism of virus-induced RIPK3 activation remains elusive. Interferon-inducible 2'-5' oligoadenylate synthetase-like (OASL) protein is devoid of enzymatic function but displays potent antiviral activity. Here we describe a role of OASL as a virus-induced necroptosis promoter that scaffolds the RIPK3-ZBP1 non-canonical necrosome via liquid-like phase condensation. This liquid-like platform of OASL recruits RIPK3 and ZBP1 via protein-protein interactions to provide spatial segregation for RIPK3 nucleation. This process facilitates the amyloid-like fibril formation and activation of RIPK3 and thereby MLKL phosphorylation for necroptosis. Mice deficient in Oasl1 exhibit severely impaired necroptosis and attenuated inflammation after viral infection, resulting in uncontrolled viral dissemination and lethality. Our study demonstrates an interferon-induced innate response whereby OASL scaffolds RIPK3-ZBP1 assembly via its phase-separated liquid droplets to facilitate necroptosis-mediated antiviral immunity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



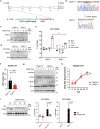
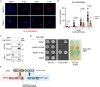

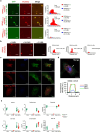
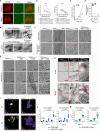

References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI171201/AI/NIAID NIH HHS/United States
- R01 AI140718/AI/NIAID NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- K99 HL153688/HL/NHLBI NIH HHS/United States
- R01 AI140705/AI/NIAID NIH HHS/United States
- R35 CA200422/CA/NCI NIH HHS/United States
- R00 DE028573/DE/NIDCR NIH HHS/United States
- R01 DE023926/DE/NIDCR NIH HHS/United States
- R01 CA251275/CA/NCI NIH HHS/United States
- U19 AI171443/AI/NIAID NIH HHS/United States
- R01 AI152190/AI/NIAID NIH HHS/United States
- R01 DE028521/DE/NIDCR NIH HHS/United States
- K99 DE028573/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

