Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation
- PMID: 36604598
- PMCID: PMC9892523
- DOI: 10.1038/s41422-022-00755-2
Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation
Abstract
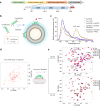
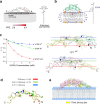
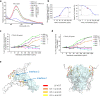
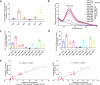
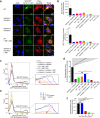
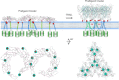
Members of the tumor necrosis factor receptor superfamily (TNFRSF) are important therapeutic targets that can be activated to induce death of cancer cells or stimulate proliferation of immune cells. Although it has long been implicated that these receptors assemble preligand associated states that are required for dominant interference in human disease, such states have so far eluded structural characterization. Here, we find that the ectodomain of death receptor 5 (DR5-ECD), a representative member of TNFRSF, can specifically self-associate when anchored to lipid bilayer, and we report this self-association structure determined by nuclear magnetic resonance (NMR). Unexpectedly, two non-overlapping interaction interfaces are identified that could propagate to higher-order clusters. Structure-guided mutagenesis indicates that the observed preligand association structure is represented on DR5-expressing cells. The DR5 preligand association serves an autoinhibitory role as single-domain antibodies (sdAbs) that partially dissociate the preligand cluster can sensitize the receptor to its ligand TRAIL and even induce substantial receptor signaling in the absence of TRAIL. Unlike most agonistic antibodies that require multivalent binding to aggregate receptors for activation, these agonistic sdAbs are monovalent and act specifically on an oligomeric, autoinhibitory configuration of the receptor. Our data indicate that receptors such as DR5 can form structurally defined preclusters incompatible with signaling and that true agonists should disrupt the preligand cluster while converting it to signaling-productive cluster. This mechanism enhances our understanding of a long-standing question in TNFRSF signaling and suggests a new opportunity for developing agonistic molecules by targeting receptor preligand clustering.
© 2022. The Author(s) under exclusive licence to Center for Excellence in Molecular Cell Science, CAS.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
Death receptor 5 rises to the occasion.Cell Res. 2023 Mar;33(3):199-200. doi: 10.1038/s41422-022-00772-1. Cell Res. 2023. PMID: 36646764 Free PMC article. No abstract available.
References
-
- Janes PW, Nievergall E, Lackmann M. Concepts and consequences of Eph receptor clustering. Semin. Cell Dev. Biol. 2012;23:43–50. - PubMed
-
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

