Netrin-1 mediates nerve innervation and angiogenesis leading to discogenic pain
- PMID: 36605621
- PMCID: PMC9804017
- DOI: 10.1016/j.jot.2022.11.006
Netrin-1 mediates nerve innervation and angiogenesis leading to discogenic pain
Abstract
Objective: Discogenic low back pain (LBP) is associated with nociceptive nerve fibers that grow into degenerated intervertebral discs (IVD) but the etiopathogenesis of disease is not fully understood. The purpose of this study was to clarify the role of Netrin-1 in causing discogenic LBP.
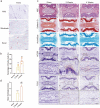
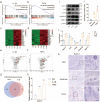
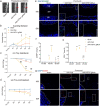
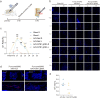
Methods: The level of nociceptive nerve innervation was examined in disc degenerative patients and rat needle-punctured models by immunohistochemistry. Nucleus pulposus (NP) cells were isolated from IVD tissues of rats and induced degeneration by interleukin-1β (IL-1β) or tumor necrosis factor α (TNFα). The candidate genes related to neuron outgrowth and migration were selected by Next-generation sequencing (NGS). CRISPR/Cas9 was used to knockdown Netrin-1 in NP cells. The impact of Netrin-1 on nerve innervation were evaluated with P2X2、NF200 staining and microfluidics assay. Meanwhile the CD31 staining and transwell assay were used to evaluate the impact of Netrin-1 in angiogenesis. The proteins and RNA extracted from NP cells related to catabolism and anabolism were examined by western blot assay and RT-qPCR experiment. ChIP and luciferase experiments were used to assess the intracellular transcriptional regulation of Netrin-1. Further, a needle-punctured rat model followed by histomorphometry and immunofluorescence histochemistry was used to explore the potential effect of Netrin-1 on LBP in vivo.
Results: The level of nerve innervation was increased in severe disc degenerative patients while the expression of Netrin-1 was upregulated. The supernatants of NP cells stimulated with IL-1β or TNFα containing more Netrin-1 could promote axon growth and vascular endothelial cells migration. Knocking down Netrin-1 or overexpressing transcription factor TCF3 as a negative regulator of Netrin-1 attenuated this effect. The needle-punctured rat model brought significant spinal hypersensitivity, nerve innervation and angiogenesis, nevertheless knocking down Netrin-1 effectively prevented disc degeneration-induced adverse impacts.
Conclusion: Discogenic LBP was induced by Netrin-1, which mediated nerve innervation and angiogenesis in disc degeneration. Knocking down Netrin-1 by CRISPR/Cas9 or negatively regulating Netrin1 by transcription factor TCF3 could alleviate spinal hypersensitivity.
The translational potential of this article: This study on Netrin-1 could provide a new target and theoretical basis for the prevention and treatment for discogenic back pain.
Keywords: Discogenic back pain; Intervertebral disc degeneration; Nerve innervation; Netrin-1.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Expression and regulation of neurotrophic and angiogenic factors during human intervertebral disc degeneration.Arthritis Res Ther. 2014 Aug 20;16(5):416. doi: 10.1186/s13075-014-0416-1. Arthritis Res Ther. 2014. PMID: 25209447 Free PMC article.
-
Increased expression of netrin-1 and its deleted in colorectal cancer receptor in human diseased lumbar intervertebral disc compared with autopsy control.Spine (Phila Pa 1976). 2012 Dec 1;37(25):2074-81. doi: 10.1097/BRS.0b013e31825d4ebc. Spine (Phila Pa 1976). 2012. PMID: 22588384
-
Stem Cell Therapies for Treatment of Discogenic Low Back Pain: a Comprehensive Review.Curr Pain Headache Rep. 2019 Jul 29;23(9):65. doi: 10.1007/s11916-019-0804-y. Curr Pain Headache Rep. 2019. PMID: 31359164 Review.
-
Pathomechanisms of discogenic low back pain in humans and animal models.Spine J. 2015 Jun 1;15(6):1347-55. doi: 10.1016/j.spinee.2013.07.490. Epub 2014 Mar 20. Spine J. 2015. PMID: 24657737 Review.
-
ISSLS prize winner: disc dynamic compression in rats produces long-lasting increases in inflammatory mediators in discs and induces long-lasting nerve injury and regeneration of the afferent fibers innervating discs: a pathomechanism for chronic discogenic low back pain.Spine (Phila Pa 1976). 2012 Oct 1;37(21):1810-8. doi: 10.1097/BRS.0b013e31824ffac6. Spine (Phila Pa 1976). 2012. PMID: 22366969
Cited by
-
Updates on Pathophysiology of Discogenic Back Pain.J Clin Med. 2023 Nov 2;12(21):6907. doi: 10.3390/jcm12216907. J Clin Med. 2023. PMID: 37959372 Free PMC article. Review.
-
Addressing musculoskeletal diseases by exploring the potentials of stem cells and plant-derived chemicals.J Orthop Translat. 2023 Apr 27;39:A1-A2. doi: 10.1016/j.jot.2023.04.004. eCollection 2023 Mar. J Orthop Translat. 2023. PMID: 37187999 Free PMC article. No abstract available.
-
Preserving the Immune-Privileged Niche of the Nucleus Pulposus: Safeguarding Intervertebral Discs from Degeneration after Discectomy with Synthetic Mucin Hydrogel Injection.Adv Sci (Weinh). 2024 Nov;11(43):e2404496. doi: 10.1002/advs.202404496. Epub 2024 Aug 29. Adv Sci (Weinh). 2024. PMID: 39207014 Free PMC article.
-
Remodeling of the Intra-Conduit Inflammatory Microenvironment to Improve Peripheral Nerve Regeneration with a Neuromechanical Matching Protein-Based Conduit.Adv Sci (Weinh). 2024 May;11(17):e2302988. doi: 10.1002/advs.202302988. Epub 2024 Mar 2. Adv Sci (Weinh). 2024. PMID: 38430538 Free PMC article.
-
Metastatic Infiltration of Nervous Tissue and Periosteal Nerve Sprouting in Multiple Myeloma-Induced Bone Pain in Mice and Human.J Neurosci. 2023 Jul 19;43(29):5414-5430. doi: 10.1523/JNEUROSCI.0404-23.2023. Epub 2023 Jun 7. J Neurosci. 2023. PMID: 37286351 Free PMC article.
References
-
- Hoy D., Brooks P., Blyth F., Buchbinder R. The Epidemiology of low back pain. Best Pract Res Clin Rheumatol. 2010;24:769–781. - PubMed
-
- Hoy D., March L., Brooks P., Blyth F., Woolf A., Bain C., et al. The global burden of low back pain: estimates from the Global Burden of Disease 2010 study. Ann Rheum Dis. 2014;73:968–974. - PubMed
-
- Katz J.N. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88(Suppl 2):21–24. - PubMed
-
- Deyo R.A., Weinstein J.N. Low back pain. N Engl J Med. 2001;344:363–370. - PubMed
-
- Urits I., Burshtein A., Sharma M., Testa L., Gold P.A., Orhurhu V., et al. Low Back Pain, a Comprehensive Review: Pathophysiology, Diagnosis, and Treatment. Curr Pain Headache Rep. 2019;23:23. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

