The role of SWI/SNF chromatin remodelers in the repair of DNA double strand breaks and cancer therapy
- PMID: 36605718
- PMCID: PMC9810387
- DOI: 10.3389/fcell.2022.1071786
The role of SWI/SNF chromatin remodelers in the repair of DNA double strand breaks and cancer therapy
Abstract
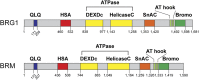
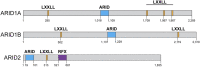
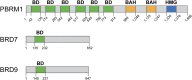
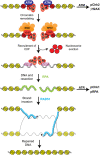
Switch/Sucrose non-fermenting (SWI/SNF) chromatin remodelers hydrolyze ATP to push and slide nucleosomes along the DNA thus modulating access to various genomic loci. These complexes are the most frequently mutated epigenetic regulators in human cancers. SWI/SNF complexes are well known for their function in transcription regulation, but more recent work has uncovered a role for these complexes in the repair of DNA double strand breaks (DSBs). As radiotherapy and most chemotherapeutic agents kill cancer cells by inducing double strand breaks, by identifying a role for these complexes in double strand break repair we are also identifying a DNA repair vulnerability that can be exploited therapeutically in the treatment of SWI/SNF-mutated cancers. In this review we summarize work describing the function of various SWI/SNF subunits in the repair of double strand breaks with a focus on homologous recombination repair and discuss the implication for the treatment of cancers with SWI/SNF mutations.
Keywords: DNA end resection; PARP inhibitors; SWI/SNF; cancer therapy; chromatin remodelers; double strand break repair; homologous recombination.
Copyright © 2022 Sadek, Sheth, Zimmerman, Hays and Vélez-Cruz.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

