Bridging physiological and perceptual views of autism by means of sampling-based Bayesian inference
- PMID: 36605888
- PMCID: PMC9810278
- DOI: 10.1162/netn_a_00219
Bridging physiological and perceptual views of autism by means of sampling-based Bayesian inference
Abstract
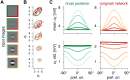
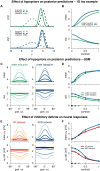
Theories for autism spectrum disorder (ASD) have been formulated at different levels, ranging from physiological observations to perceptual and behavioral descriptions. Understanding the physiological underpinnings of perceptual traits in ASD remains a significant challenge in the field. Here we show how a recurrent neural circuit model that was optimized to perform sampling-based inference and displays characteristic features of cortical dynamics can help bridge this gap. The model was able to establish a mechanistic link between two descriptive levels for ASD: a physiological level, in terms of inhibitory dysfunction, neural variability, and oscillations, and a perceptual level, in terms of hypopriors in Bayesian computations. We took two parallel paths-inducing hypopriors in the probabilistic model, and an inhibitory dysfunction in the network model-which lead to consistent results in terms of the represented posteriors, providing support for the view that both descriptions might constitute two sides of the same coin.
Keywords: Autism; Hypopriors; Inhibitory dysfunction; Neural circuits; Sampling-based inference.
© 2021 Massachusetts Institute of Technology.
Figures


References
-
- American Psychiatric Association. (2013). Diagnostic and statistical manual of mental disorders (DSM-5). American Psychiatric Association Publishing. 10.1176/appi.books.9780890425596 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous
