Dysregulation of Grainyhead-like 3 expression causes widespread developmental defects
- PMID: 36606449
- PMCID: PMC10952483
- DOI: 10.1002/dvdy.565
Dysregulation of Grainyhead-like 3 expression causes widespread developmental defects
Abstract
Background: The gene encoding the transcription factor, Grainyhead-like 3 (Grhl3), plays critical roles in mammalian development and homeostasis. Grhl3-null embryos exhibit thoraco-lumbo-sacral spina bifida and soft-tissue syndactyly. Additional studies reveal that these embryos also exhibit an epidermal proliferation/differentiation imbalance. This manifests as skin barrier defects resulting in peri-natal lethality and defective wound repair. Despite these extensive analyses of Grhl3 loss-of-function models, the consequences of gain-of-function of this gene have been difficult to achieve.
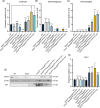
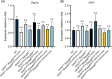
Results: In this study, we generated a novel mouse model that expresses Grhl3 from a transgene integrated in the Rosa26 locus on an endogenous Grhl3-null background. Expression of the transgene rescues both the neurulation and skin barrier defects of the knockout mice, allowing survival into adulthood. Despite this, the mice are not normal, exhibiting a range of phenotypes attributable to dysregulated Grhl3 expression. In mice homozygous for the transgene, we observe a severe Shaker-Waltzer phenotype associated with hearing impairment. Micro-CT scanning of the inner ear revealed profound structural alterations underlying these phenotypes. In addition, these mice exhibit other developmental anomalies including hair loss, digit defects, and epidermal dysmorphogenesis.
Conclusion: Taken together, these findings indicate that diverse developmental processes display low tolerance to dysregulation of Grhl3.
Keywords: Grhl3 overexpression; Shaker-Waltzer phenotype; inner ear malformation; neural tube defects; skin barrier defects.
© 2022 The Authors. Developmental Dynamics published by Wiley Periodicals LLC on behalf of American Association for Anatomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Wilanowski T, Tuckfield A, Cerruti L, et al. A highly conserved novel family of mammalian developmental transcription factors related to drosophila grainyhead. Mech Dev. 2002;114(1–2):37‐50. - PubMed
-
- Ting SB, Wilanowski T, Cerruti L, Zhao LL, Cunningham JM, Jane SM. The identification and characterization of human sister‐of‐mammalian Grainyhead (SOM) expands the grainyhead‐like family of developmental transcription factors. Biochem J. 2003;370(Pt 3):953‐962. doi:10.1042/bj20021476 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

