One dimensional approximations of neuronal dynamics reveal computational strategy
- PMID: 36607933
- PMCID: PMC9821456
- DOI: 10.1371/journal.pcbi.1010784
One dimensional approximations of neuronal dynamics reveal computational strategy
Abstract
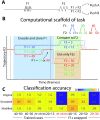
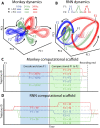
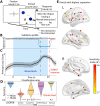
The relationship between neuronal activity and computations embodied by it remains an open question. We develop a novel methodology that condenses observed neuronal activity into a quantitatively accurate, simple, and interpretable model and validate it on diverse systems and scales from single neurons in C. elegans to fMRI in humans. The model treats neuronal activity as collections of interlocking 1-dimensional trajectories. Despite their simplicity, these models accurately predict future neuronal activity and future decisions made by human participants. Moreover, the structure formed by interconnected trajectories-a scaffold-is closely related to the computational strategy of the system. We use these scaffolds to compare the computational strategy of primates and artificial systems trained on the same task to identify specific conditions under which the artificial agent learns the same strategy as the primate. The computational strategy extracted using our methodology predicts specific errors on novel stimuli. These results show that our methodology is a powerful tool for studying the relationship between computation and neuronal activity across diverse systems.
Copyright: © 2023 Brennan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

