A proteomic perspective and involvement of cytokines in SARS-CoV-2 infection
- PMID: 36608055
- PMCID: PMC9821788
- DOI: 10.1371/journal.pone.0279998
A proteomic perspective and involvement of cytokines in SARS-CoV-2 infection
Abstract
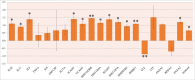
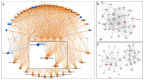
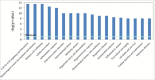
Infection with the SARS-CoV-2 virus results in manifestation of several clinical observations from asymptomatic to multi-organ failure. Biochemically, the serious effects are due to what is described as cytokine storm. The initial infection region for COVID-19 is the nasopharyngeal/oropharyngeal region which is the site where samples are taken to examine the presence of virus. We have now carried out detailed proteomic analysis of the nasopharyngeal/oropharyngeal swab samples collected from normal individuals and those tested positive for SARS-CoV-2, in India, during the early days of the pandemic in 2020, by RTPCR, involving high throughput quantitative proteomics analysis. Several proteins like annexins, cytokines and histones were found differentially regulated in the host human cells following SARS-CoV-2 infection. Genes for these proteins were also observed to be differentially regulated when their expression was analyzed. Majority of the cytokine proteins were found to be up regulated in the infected individuals. Cell to Cell signaling interaction, Immune cell trafficking and inflammatory response pathways were found associated with the differentially regulated proteins based on network pathway analysis.
Copyright: © 2023 Banu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

