Pathogenic tau-induced transposable element-derived dsRNA drives neuroinflammation
- PMID: 36608133
- PMCID: PMC9821943
- DOI: 10.1126/sciadv.abq5423
Pathogenic tau-induced transposable element-derived dsRNA drives neuroinflammation
Abstract
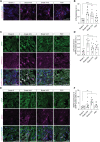
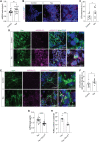
Deposition of tau protein aggregates in the brain of affected individuals is a defining feature of "tauopathies," including Alzheimer's disease. Studies of human brain tissue and various model systems of tauopathy report that toxic forms of tau negatively affect nuclear and genomic architecture, identifying pathogenic tau-induced heterochromatin decondensation and consequent retrotransposon activation as a causal mediator of neurodegeneration. On the basis of their similarity to retroviruses, retrotransposons drive neuroinflammation via toxic intermediates, including double-stranded RNA (dsRNA). We find that dsRNA and dsRNA sensing machinery are elevated in astrocytes of postmortem brain tissue from patients with Alzheimer's disease and progressive supranuclear palsy and in brains of tau transgenic mice. Using a Drosophila model of tauopathy, we identify specific tau-induced retrotransposons that form dsRNA and find that pathogenic tau and heterochromatin decondensation causally drive dsRNA-mediated neurodegeneration and neuroinflammation. Our study suggests that pathogenic tau-induced heterochromatin decondensation and retrotransposon activation cause elevation of inflammatory, transposable element-derived dsRNA in the adult brain.
Figures





References
-
- V. Gorbunova, A. Seluanov, P. Mita, W. McKerrow, D. Fenyö, J. D. Boeke, S. B. Linker, F. H. Gage, J. A. Kreiling, A. P. Petrashen, T. A. Woodham, J. R. Taylor, S. L. Helfand, J. M. Sedivy, The role of retrotransposable elements in ageing and age-associated diseases. Nature 596, 43–53 (2021). - PMC - PubMed
-
- S. Lanciano, G. Cristofari, Measuring and interpreting transposable element expression. Nat. Rev. Genet. 21, 721–736 (2020). - PubMed
-
- H. Kaneko, S. Dridi, V. Tarallo, B. D. Gelfand, B. J. Fowler, W. G. Cho, M. E. Kleinman, S. L. Ponicsan, W. W. Hauswirth, V. A. Chiodo, K. Karikó, J. W. Yoo, D. K. Lee, M. Hadziahmetovic, Y. Song, S. Misra, G. Chaudhuri, F. W. Buaas, R. E. Braun, D. R. Hinton, Q. Zhang, H. E. Grossniklaus, J. M. Provis, M. C. Madigan, A. H. Milam, N. L. Justice, R. J. C. Albuquerque, A. D. Blandford, S. Bogdanovich, Y. Hirano, J. Witta, E. Fuchs, D. R. Littman, B. K. Ambati, C. M. Rudin, M. M. W. Chong, P. Provost, J. F. Kugel, J. A. Goodrich, J. L. Dunaief, J. Z. Baffi, J. Ambati, DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471, 325–332 (2011). - PMC - PubMed
-
- P. Ramirez, G. Zuniga, W. Sun, A. Beckmann, E. Ochoa, S. L. DeVos, B. Hyman, G. Chiu, E. R. Roy, W. Cao, M. Orr, V. Buggia-Prevot, W. J. Ray, B. Frost, Pathogenic tau accelerates aging-associated activation of transposable elements in the mouse central nervous system. Prog. Neurobiol. 208, 102181 (2022). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

