Epithelial-intrinsic defects in TGFβR signaling drive local allergic inflammation manifesting as eosinophilic esophagitis
- PMID: 36608150
- PMCID: PMC10106118
- DOI: 10.1126/sciimmunol.abp9940
Epithelial-intrinsic defects in TGFβR signaling drive local allergic inflammation manifesting as eosinophilic esophagitis
Abstract
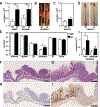
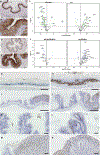
Allergic diseases are a global health challenge. Individuals harboring loss-of-function variants in transforming growth factor-β receptor (TGFβR) genes have an increased prevalence of allergic disorders, including eosinophilic esophagitis. Allergic diseases typically localize to mucosal barriers, implicating epithelial dysfunction as a cardinal feature of allergic disease. Here, we describe an essential role for TGFβ in the control of tissue-specific immune homeostasis that provides mechanistic insight into these clinical associations. Mice expressing a TGFβR1 loss-of-function variant identified in atopic patients spontaneously develop disease that clinically, immunologically, histologically, and transcriptionally recapitulates eosinophilic esophagitis. In vivo and in vitro, TGFβR1 variant-expressing epithelial cells are hyperproliferative, fail to differentiate properly, and overexpress innate proinflammatory mediators, which persist in the absence of lymphocytes or external allergens. Together, our results support the concept that TGFβ plays a fundamental, nonredundant, epithelial cell-intrinsic role in controlling tissue-specific allergic inflammation that is independent of its role in adaptive immunity.
Conflict of interest statement
Figures






References
-
- Cardoso S, Robertson SP, Daniel PB, TGFBR1 mutations associated with Loeys-Dietz syndrome are inactivating. J Recept Signal Transduct Res 32, 150–155 (2012). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

