Microfluidic Platform for Profiling of Extracellular Vesicles from Single Breast Cancer Cells
- PMID: 36608325
- PMCID: PMC9878503
- DOI: 10.1021/acs.analchem.2c04106
Microfluidic Platform for Profiling of Extracellular Vesicles from Single Breast Cancer Cells
Abstract
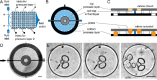
Extracellular vesicles (EVs) are considered as valuable biomarkers to discriminate healthy from diseased cells such as cancer. Passing cytosolic and plasma membranes before their release, EVs inherit the biochemical properties of the cell. Here, we determine protein profiles of single EVs to understand how much they represent their cell of origin. We use a microfluidic platform which allows to immobilize EVs from completely isolated single cells, reducing heterogeneity of EVs as strongly seen in cell populations. After immunostaining, we employ four-color total internal reflection fluorescence microscopy to enumerate EVs and determine their biochemical fingerprint encoded in membranous or cytosolic proteins. Analyzing single cells derived from pleural effusions of two different human adenocarcinoma as well as from human embryonic kidney (SkBr3, MCF-7 and HEK293, respectively), we observed that a single cell secretes enough EVs to extract the respective tissue fingerprint. We show that overexpressed integral plasma membrane proteins are also found in EV membranes, which together with populations of colocalized proteins, provide a cell-specific, characteristic pattern. Our method highlights the potential of EVs as a diagnostic marker and can be directly employed for fundamental studies of EV biogenesis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Ford C. H. J.; Maie A. L. B.; Bushra A. L. A.; Issam F. Reassessment of Estrogen Receptor Expression in Human Breast Cancer Cell Lines. Anticancer Res. 2011, 31 (2), 521–527. - PubMed
LinkOut - more resources
Full Text Sources

