Phage therapy: From biological mechanisms to future directions
- PMID: 36608652
- PMCID: PMC9827498
- DOI: 10.1016/j.cell.2022.11.017
Phage therapy: From biological mechanisms to future directions
Abstract
Increasing antimicrobial resistance rates have revitalized bacteriophage (phage) research, the natural predators of bacteria discovered over 100 years ago. In order to use phages therapeutically, they should (1) preferably be lytic, (2) kill the bacterial host efficiently, and (3) be fully characterized to exclude side effects. Developing therapeutic phages takes a coordinated effort of multiple stakeholders. Herein, we review the state of the art in phage therapy, covering biological mechanisms, clinical applications, remaining challenges, and future directions involving naturally occurring and genetically modified or synthetic phages.
Keywords: bacteriophage; bacteriophage therapy; phage; phage therapy.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.A.S. owns stock in Adaptive Phage Therapeutics and is an unpaid advisor to Felix Biosciences. R.T.S. is a scientific consultant to LyseNTech and GSK. G.F.H. receives support from Janssen Inc through a Collaborative Research Agreement and is a consultant for Janssen, Inc. and Tessera, Inc. V.K.M. is a co-founder of Felix Biotechnology.
Figures


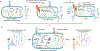

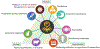
References
-
- World Health Organiztion (2017). WHO publishes list of bacteria for which new antibiotics are urgently needed. In Davies OL, ed. News release. WHO.
-
- CDC (2022). COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022. 10.15620/cdc:117915. - DOI
-
- CDC (2022). COVID-19 & Antimicrobial Resistance. https://www.cdc.gov/drugresistance/covid19.html.
-
- O’Neill J (2016). Tackling drug-resistant infections globally: final report and recommendations. Report Government of the United Kingdom. https://apo.org.au/node/63983.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

