Enhanced production of D-pantothenic acid in Corynebacterium glutamicum using an efficient CRISPR-Cpf1 genome editing method
- PMID: 36609377
- PMCID: PMC9817396
- DOI: 10.1186/s12934-023-02017-1
Enhanced production of D-pantothenic acid in Corynebacterium glutamicum using an efficient CRISPR-Cpf1 genome editing method
Abstract
Background: Corynebacterium glutamicum has industrial track records for producing a variety of valuable products such as amino acids. Although CRISPR-based genome editing technologies have undergone immense developments in recent years, the suicide-plasmid-based approaches are still predominant for C. glutamicum genome manipulation. It is crucial to develop a simple and efficient CRISPR genome editing method for C. glutamicum.
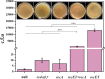
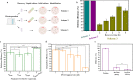
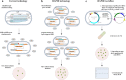
Results: In this study, we developed a RecombinAtion Prior to Induced Double-strand-break (RAPID) genome editing technology for C. glutamicum, as Cpf1 cleavage was found to disrupt RecET-mediated homologous recombination (HR) of the donor template into the genome. The RAPID toolbox enabled highly efficient gene deletion and insertion, and notably, a linear DNA template was sufficient for gene deletion. Due to the simplified procedure and iterative operation ability, this methodology could be widely applied in C. glutamicum genetic manipulations. As a proof of concept, a high-yield D-pantothenic acid (vitamin B5)-producing strain was constructed, which, to the best of our knowledge, achieved the highest reported titer of 18.62 g/L from glucose only.
Conclusions: We developed a RecET-assisted CRISPR-Cpf1 genome editing technology for C. glutamicum that harnessed CRISPR-induced DSBs as a counterselection. This method is of great importance to C. glutamicum genome editing in terms of its practical applications, which also guides the development of CRISPR genome editing tools for other microorganisms.
Keywords: CRISPR-Cpf1; Corynebacterium glutamicum; Genome editing; Metabolic engineering; Synthetic biology; Vitamin.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Kallscheuer N, Vogt M, Stenzel A, Gätgens J, Bott M, Marienhagen J. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Meta Eng. 2016;38:47–55. - PubMed
-
- Lee SY, Kim HU. Systems strategies for developing industrial microbial strains. Nat Biotechnol. 2015;33:1061–1072. - PubMed
-
- Gordillo Sierra AR, Alper HS. Progress in the metabolic engineering of bio-based lactams and their ω-amino acids precursors. Biotechnol Adv. 2020;43:107587. - PubMed
-
- Becker J, Gießelmann G, Hoffmann SL, Wittmann C. Corynebacterium glutamicum for sustainable bioproduction: from metabolic physiology to systems metabolic engineering. Adv Biochem Eng Biotechnol. 2018;162:217–263. - PubMed
-
- Becker J, Rohles CM, Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab Eng. 2018;50:122–141. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

