Molecular subtypes of ALS are associated with differences in patient prognosis
- PMID: 36609402
- PMCID: PMC9822908
- DOI: 10.1038/s41467-022-35494-w
Molecular subtypes of ALS are associated with differences in patient prognosis
Abstract
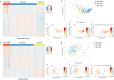
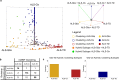
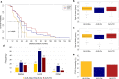
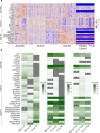
Amyotrophic Lateral Sclerosis (ALS) is a neurodegenerative disease with poorly understood clinical heterogeneity, underscored by significant differences in patient age at onset, symptom progression, therapeutic response, disease duration, and comorbidity presentation. We perform a patient stratification analysis to better understand the variability in ALS pathology, utilizing postmortem frontal and motor cortex transcriptomes derived from 208 patients. Building on the emerging role of transposable element (TE) expression in ALS, we consider locus-specific TEs as distinct molecular features during stratification. Here, we identify three unique molecular subtypes in this ALS cohort, with significant differences in patient survival. These results suggest independent disease mechanisms drive some of the clinical heterogeneity in ALS.
© 2023. The Author(s).
Conflict of interest statement
R.B. is the chairman of the board of Iron Horse Diagnostics, which has not contributed financially, or by any other means, to this study. The remaining authors declare no competing interests.
Figures


References
-
- Mitchell JD, et al. Timelines in the diagnostic evaluation of people with suspected amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND)–a 20-year review: can we do better? Amyotroph. Lateral Scler. 2010;11:537–541. - PubMed
-
- Bowser R, Turner MR, Shefner J. Biomarkers in amyotrophic lateral sclerosis: opportunities and limitations. Nat. Rev. Neurol. 2011;7:631–638. - PubMed
-
- Calvo A, et al. Factors predicting survival in ALS: a multicenter Italian study. J. Neurol. 2017;264:54–63. - PubMed
-
- Walhout, R., Verstraete, E., Van Den Heuvel, M. P., Veldink, J. H. & Van Den Berg, L.H. Patterns of symptom development in patients with motor neuron disease. Amyotroph. Lateral Scler. Frontotemporal Degener.19, 21–28 (2018). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

