P-TarPmiR accurately predicts plant-specific miRNA targets
- PMID: 36609461
- PMCID: PMC9822942
- DOI: 10.1038/s41598-022-27283-8
P-TarPmiR accurately predicts plant-specific miRNA targets
Abstract
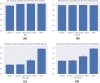
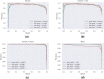
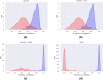
microRNAs (miRNAs) are small non-coding ribonucleic acids that post-transcriptionally regulate gene expression through the targeting of messenger RNA (mRNAs). Most miRNA target predictors have focused on animal species and prediction performance drops substantially when applied to plant species. Several rule-based miRNA target predictors have been developed in plant species, but they often fail to discover new miRNA targets with non-canonical miRNA-mRNA binding. Here, the recently published TarDB database of plant miRNA-mRNA data is leveraged to retrain the TarPmiR miRNA target predictor for application on plant species. Rigorous experiment design across four plant test species demonstrates that animal-trained predictors fail to sustain performance on plant species, and that the use of plant-specific training data improves accuracy depending on the quantity of plant training data used. Surprisingly, our results indicate that the complete exclusion of animal training data leads to the most accurate plant-specific miRNA target predictor indicating that animal-based data may detract from miRNA target prediction in plants. Our final plant-specific miRNA prediction method, dubbed P-TarPmiR, is freely available for use at http://ptarpmir.cu-bic.ca . The final P-TarPmiR method is used to predict targets for all miRNA within the soybean genome. Those ranked predictions, together with GO term enrichment, are shared with the research community.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. - PubMed
-
- Dai X, Zhuang Z, Zhao PX. Computational analysis of mirna targets in plants: Current status and challenges. Brief. Bioinform. 2011;12:115–121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

