STING agonism overcomes STAT3-mediated immunosuppression and adaptive resistance to PARP inhibition in ovarian cancer
- PMID: 36609487
- PMCID: PMC9827255
- DOI: 10.1136/jitc-2022-005627
STING agonism overcomes STAT3-mediated immunosuppression and adaptive resistance to PARP inhibition in ovarian cancer
Abstract
Background: Poly (ADP-ribose) polymerase (PARP) inhibition (PARPi) has demonstrated potent therapeutic efficacy in patients with BRCA-mutant ovarian cancer. However, acquired resistance to PARPi remains a major challenge in the clinic.
Methods: PARPi-resistant ovarian cancer mouse models were generated by long-term treatment of olaparib in syngeneic Brca1-deficient ovarian tumors. Signal transducer and activator of transcription 3 (STAT3)-mediated immunosuppression was investigated in vitro by co-culture experiments and in vivo by analysis of immune cells in the tumor microenvironment (TME) of human and mouse PARPi-resistant tumors. Whole genome transcriptome analysis was performed to assess the antitumor immunomodulatory effect of STING (stimulator of interferon genes) agonists on myeloid cells in the TME of PARPi-resistant ovarian tumors. A STING agonist was used to overcome STAT3-mediated immunosuppression and acquired PARPi resistance in syngeneic and patient-derived xenografts models of ovarian cancer.
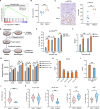
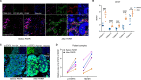
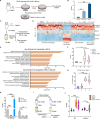
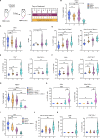
Results: In this study, we uncover an adaptive resistance mechanism to PARP inhibition mediated by tumor-associated macrophages (TAMs) in the TME. Markedly increased populations of protumor macrophages are found in BRCA-deficient ovarian tumors that rendered resistance to PARPi in both murine models and patients. Mechanistically, PARP inhibition elevates the STAT3 signaling pathway in tumor cells, which in turn promotes protumor polarization of TAMs. STAT3 ablation in tumor cells mitigates polarization of protumor macrophages and increases tumor-infiltrating T cells on PARP inhibition. These findings are corroborated in patient-derived, PARPi-resistant BRCA1-mutant ovarian tumors. Importantly, STING agonists reshape the immunosuppressive TME by reprogramming myeloid cells and overcome the TME-dependent adaptive resistance to PARPi in ovarian cancer. This effect is further enhanced by addition of the programmed cell death protein-1 blockade.
Conclusions: We elucidate an adaptive immunosuppression mechanism rendering resistance to PARPi in BRCA1-mutant ovarian tumors. This is mediated by enrichment of protumor TAMs propelled by PARPi-induced STAT3 activation in tumor cells. We also provide a new strategy to reshape the immunosuppressive TME with STING agonists and overcome PARPi resistance in ovarian cancer.
Keywords: Drug Therapy, Combination; Immunomodulation; Immunotherapy; Macrophages; Tumor Microenvironment.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: LD, QW and JJZ are co-inventors of the patent related to this study (US application 63/148426). QW is a scientific consultant for Crimson Biopharm. PAK is a scientific consultant for Alkermes, AstraZeneca, Bayer, GSK, Merck, Pfizer, Tesaro, Mersana, Repare, Kadmon and served as the principal investigator (PI) of institutional support clinical trial for AstraZeneca, Bayer, GSK, Merck, Lilly, and BMS. TMR is cofounder of Crimson Biopharm and Geode Therapeutics, and a SAB member for Shiftbio and K2B Therapeutics. GJF has patents/pending royalties on the PD-L1/PD-1 pathway from Roche, Merck MSD, Bristol Myers Squibb, Merck KGA, Boehringer Ingelheim, AstraZeneca, Dako, Leica, Mayo Clinic, Eli Lilly, and Novartis. GJF has served on advisory boards for Roche, Bristol Myers Squibb, Origimed, Triursus, iTeos, NextPoint, IgM, Jubilant, Trillium, GV20, IOME, and Geode. GJF has equity in Nextpoint, Triursus, Xios, iTeos, IgM, Trillium, Invaria, GV20, and Geode. JFL has served as a scientific consultant for Genentech/Roche and Bristol Myers Squibb, and has been on advisory boards for Clovis Oncology, Genentech/Roche, GlaxoSmithKline, Regeneron, AstraZeneca, and Eisai. JFL has served as the institutional PI (support to the institution) of clinical trials by Genentech/Roche, AstraZeneca, Boston Biomedical, Acetylon Pharmaceuticals, Bristol Myers Squibb, Agenus, CytomX Therapeutics, Regeneron, Tesaro/GSK, Clovis Oncology, Surface Oncology, 2X Oncology, Vigeo Therapeutics, Aravive, Arch Oncology and Zentalis Pharmaceuticals. UM has served on the advisory boards of 2X Oncology, Fujifilm, Immunogen, Mersana, Geneos, and Merck. JJZ is cofounder and director of Crimson Biopharm and Geode Therapeutics. The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
