COVID-19-associated monocytic encephalitis (CAME): histological and proteomic evidence from autopsy
- PMID: 36609561
- PMCID: PMC9816522
- DOI: 10.1038/s41392-022-01291-6
COVID-19-associated monocytic encephalitis (CAME): histological and proteomic evidence from autopsy
Abstract
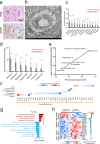
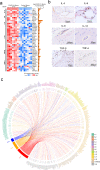
Severe neurological symptoms are associated with Coronavirus disease 2019 (COVID-19). However, the morphologic features, pathological nature and their potential mechanisms in patient brains have not been revealed despite evidence of neurotropic infection. In this study, neuropathological damages and infiltrating inflammatory cells were quantitatively evaluated by immunohistochemical staining, ultrastructural examination under electron microscopy, and an image threshold method, in postmortem brains from nine critically ill COVID-19 patients and nine age-matched cadavers of healthy individuals. Differentially expressed proteins were identified by quantitative proteomic assays. Histopathological findings included neurophagocytosis, microglia nodules, satellite phenomena, extensive edema, focal hemorrhage, and infarction, as well as infiltrating mononuclear cells. Immunostaining of COVID-19 brains revealed extensive activation of both microglia and astrocytes, severe damage of the blood-brain barrier (BBB) and various degrees of perivascular infiltration by predominantly CD14+/CD16+/CD141+/CCR7+/CD11c+ monocytes and occasionally CD4+/CD8+ T lymphocytes. Quantitative proteomic assays combined with bioinformatics analysis identified upregulated proteins predominantly involved in immune responses, autophagy and cellular metabolism in COVID-19 patient brains compared with control brains. Proteins involved in brain development, neuroprotection, and extracellular matrix proteins of the basement membrane were downregulated, potentially caused by the activation of transforming growth factor β receptor and vascular endothelial growth factor signaling pathways. Thus, our results define histopathological and molecular profiles of COVID-19-associated monocytic encephalitis (CAME) and suggest potential therapeutic targets.
© 2023. The Author(s).
Conflict of interest statement
X.-W.B. is the editorial board member of Signal Transduction and Targeted Therapy, but he has not been involved in the process of manuscript handling.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

