HSP70-binding motifs function as protein quality control degrons
- PMID: 36609589
- PMCID: PMC11072582
- DOI: 10.1007/s00018-022-04679-3
HSP70-binding motifs function as protein quality control degrons
Abstract
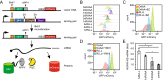
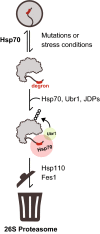
Protein quality control (PQC) degrons are short protein segments that target misfolded proteins for proteasomal degradation, and thus protect cells against the accumulation of potentially toxic non-native proteins. Studies have shown that PQC degrons are hydrophobic and rarely contain negatively charged residues, features which are shared with chaperone-binding regions. Here we explore the notion that chaperone-binding regions may function as PQC degrons. When directly tested, we found that a canonical Hsp70-binding motif (the APPY peptide) functioned as a dose-dependent PQC degron both in yeast and in human cells. In yeast, Hsp70, Hsp110, Fes1, and the E3 Ubr1 target the APPY degron. Screening revealed that the sequence space within the chaperone-binding region of APPY that is compatible with degron function is vast. We find that the number of exposed Hsp70-binding sites in the yeast proteome correlates with a reduced protein abundance and half-life. Our results suggest that when protein folding fails, chaperone-binding sites may operate as PQC degrons, and that the sequence properties leading to PQC-linked degradation therefore overlap with those of chaperone binding.
Keywords: Chaperone; Proteasome; Protein degradation; Protein quality control; Protein stability; Protein unfolding.
© 2023. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
No conflicting interests to declare.
Figures


Similar articles
-
Degradation Signals for Ubiquitin-Proteasome Dependent Cytosolic Protein Quality Control (CytoQC) in Yeast.G3 (Bethesda). 2016 Jul 7;6(7):1853-66. doi: 10.1534/g3.116.027953. G3 (Bethesda). 2016. PMID: 27172186 Free PMC article.
-
The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein.PLoS One. 2013;8(1):e52099. doi: 10.1371/journal.pone.0052099. Epub 2013 Jan 16. PLoS One. 2013. PMID: 23341891 Free PMC article.
-
The extent of Ssa1/Ssa2 Hsp70 chaperone involvement in nuclear protein quality control degradation varies with the substrate.Mol Biol Cell. 2020 Feb 1;31(3):221-233. doi: 10.1091/mbc.E18-02-0121. Epub 2019 Dec 11. Mol Biol Cell. 2020. PMID: 31825716 Free PMC article.
-
Co-Chaperones in Targeting and Delivery of Misfolded Proteins to the 26S Proteasome.Biomolecules. 2020 Aug 4;10(8):1141. doi: 10.3390/biom10081141. Biomolecules. 2020. PMID: 32759676 Free PMC article. Review.
-
UPS-dependent strategies of protein quality control degradation.Trends Biochem Sci. 2024 Oct;49(10):859-874. doi: 10.1016/j.tibs.2024.06.006. Epub 2024 Jun 29. Trends Biochem Sci. 2024. PMID: 38945729 Review.
Cited by
-
Reversible protein assemblies in the proteostasis network in health and disease.Front Mol Biosci. 2023 Mar 20;10:1155521. doi: 10.3389/fmolb.2023.1155521. eCollection 2023. Front Mol Biosci. 2023. PMID: 37021114 Free PMC article. Review.
-
A complete map of human cytosolic degrons and their relevance for disease.bioRxiv [Preprint]. 2025 May 15:2025.05.10.653233. doi: 10.1101/2025.05.10.653233. bioRxiv. 2025. PMID: 40463067 Free PMC article. Preprint.
-
Deep mutational scanning reveals a correlation between degradation and toxicity of thousands of aspartoacylase variants.Nat Commun. 2024 May 13;15(1):4026. doi: 10.1038/s41467-024-48481-0. Nat Commun. 2024. PMID: 38740822 Free PMC article.
-
J-domain proteins: From molecular mechanisms to diseases.Cell Stress Chaperones. 2024 Feb;29(1):21-33. doi: 10.1016/j.cstres.2023.12.002. Epub 2023 Dec 23. Cell Stress Chaperones. 2024. PMID: 38320449 Free PMC article.
-
A mutational atlas for Parkin proteostasis.Nat Commun. 2024 Feb 20;15(1):1541. doi: 10.1038/s41467-024-45829-4. Nat Commun. 2024. PMID: 38378758 Free PMC article.
References
-
- Abildgaard AB, Stein A, Nielsen SV, Schultz-Knudsen K, Papaleo E, Shrikhande A, Hoffmann ER, Bernstein I, Gerdes A-M, Takahashi M, Ishioka C, Lindorff-Larsen K, Hartmann-Petersen R. Computational and cellular studies reveal structural destabilization and degradation of MLH1 variants in Lynch syndrome. Elife. 2019;8:e49138. doi: 10.7554/eLife.49138. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- NNF18OC0033950/Novo Nordisk Fonden
- NNF18OC0033950/Novo Nordisk Fonden
- NNF18OC0033950/Novo Nordisk Fonden
- NNF18OC0033926/Novo Nordisk Fonden
- NNF18OC0052441/Novo Nordisk Fonden
- NNF0071057/Novo Nordisk Fonden
- R249-2017-510/Lundbeckfonden
- R272-2017-4528/Lundbeckfonden
- 10.46540/2032-00007B &/Natur og Univers, Det Frie Forskningsråd
- 7014-00039B/Natur og Univers, Det Frie Forskningsråd
- 40526/Villum Fonden
- MIAMi/Horizon 2020 Framework Programme
- No. 722287/Horizon 2020 Framework Programme
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

