Dnmt1/Tet2-mediated changes in Cmip methylation regulate the development of nonalcoholic fatty liver disease by controlling the Gbp2-Pparγ-CD36 axis
- PMID: 36609599
- PMCID: PMC9898513
- DOI: 10.1038/s12276-022-00919-5
Dnmt1/Tet2-mediated changes in Cmip methylation regulate the development of nonalcoholic fatty liver disease by controlling the Gbp2-Pparγ-CD36 axis
Abstract
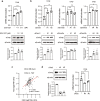
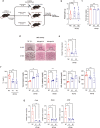
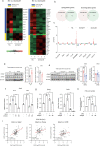
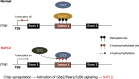
Dynamic alteration of DNA methylation leads to various human diseases, including nonalcoholic fatty liver disease (NAFLD). Although C-Maf-inducing protein (Cmip) has been reported to be associated with NAFLD, its exact underlying mechanism remains unclear. Here, we aimed to elucidate this mechanism in NAFLD in vitro and in vivo. We first identified alterations in the methylation status of the Cmip intron 1 region in mouse liver tissues with high-fat high-sucrose diet-induced NAFLD. Knockdown of DNA methyltransferase (Dnmt) 1 significantly increased Cmip expression. Chromatin immunoprecipitation assays of AML12 cells treated with oleic and palmitic acid (OPA) revealed that Dnmt1 was dissociated and that methylation of H3K27me3 was significantly decreased in the Cmip intron 1 region. Conversely, the knockdown of Tet methylcytosine dioxygenase 2 (Tet2) decreased Cmip expression. Following OPA treatment, the CCCTC-binding factor (Ctcf) was recruited, and H3K4me3 was significantly hypermethylated. Intravenous Cmip siRNA injection ameliorated NAFLD pathogenic features in ob/ob mice. Additionally, Pparγ and Cd36 expression levels were dramatically decreased in the livers of ob/ob mice administered siCmip, and RNA sequencing revealed that Gbp2 was involved. Gbp2 knockdown also induced a decrease in Pparγ and Cd36 expression, resulting in the abrogation of fatty acid uptake into cells. Our data demonstrate that Cmip and Gbp2 expression levels are enhanced in human liver tissues bearing NAFLD features. We also show that Dnmt1-Trt2/Ctcf-mediated reversible modulation of Cmip methylation regulates the Gbp2-Pparγ-Cd36 signaling pathway, indicating the potential of Cmip as a novel therapeutic target for NAFLD.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Ahrens M, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. - PubMed
-
- Botello-Manilla AE, Chavez-Tapia NC, Uribe M, Nuno-Lambarri N. Genetics and epigenetics purpose in nonalcoholic fatty liver disease. Expert. Rev. Gastroenterol. Hepatol. 2020;14:733–748. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

