Dysregulation of PRMT5 in chronic lymphocytic leukemia promotes progression with high risk of Richter's transformation
- PMID: 36609611
- PMCID: PMC9823097
- DOI: 10.1038/s41467-022-35778-1
Dysregulation of PRMT5 in chronic lymphocytic leukemia promotes progression with high risk of Richter's transformation
Abstract
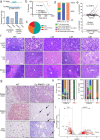
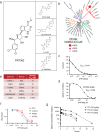
Richter's Transformation (RT) is a poorly understood and fatal progression of chronic lymphocytic leukemia (CLL) manifesting histologically as diffuse large B-cell lymphoma. Protein arginine methyltransferase 5 (PRMT5) is implicated in lymphomagenesis, but its role in CLL or RT progression is unknown. We demonstrate herein that tumors uniformly overexpress PRMT5 in patients with progression to RT. Furthermore, mice with B-specific overexpression of hPRMT5 develop a B-lymphoid expansion with increased risk of death, and Eµ-PRMT5/TCL1 double transgenic mice develop a highly aggressive disease with transformation that histologically resembles RT; where large-scale transcriptional profiling identifies oncogenic pathways mediating PRMT5-driven disease progression. Lastly, we report the development of a SAM-competitive PRMT5 inhibitor, PRT382, with exclusive selectivity and optimal in vitro and in vivo activity compared to available PRMT5 inhibitors. Taken together, the discovery that PRMT5 drives oncogenic pathways promoting RT provides a compelling rationale for clinical investigation of PRMT5 inhibitors such as PRT382 in aggressive CLL/RT cases.
© 2023. The Author(s).
Conflict of interest statement
R.B. has received research funding from Prelude Therapeutics. P.S., M.W., P.P., C.X., and K.V. are employees of Prelude Therapeutics. All the remaining authors declare no competing interests. This does not alter our adherence to
Figures





References
-
- Anderson MA, et al. Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood. 2017;129:3362–3370. - PubMed
-
- Rinaldi A, et al. Promoter methylation patterns in Richter syndrome affect stem-cell maintenance and cell cycle regulation and differ from de novo diffuse large B-cell lymphoma. Br. J. Haematol. 2013;163:194–204. - PubMed
-
- Rossi D, et al. Molecular history of Richter syndrome: origin from a cell already present at the time of chronic lymphocytic leukemia diagnosis. Int. J. Cancer J. Int. du cancer. 2012;130:3006–3010. - PubMed
-
- Rossi D, et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood. 2011;117:3391–3401. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

