Brucella effectors NyxA and NyxB target SENP3 to modulate the subcellular localisation of nucleolar proteins
- PMID: 36609656
- PMCID: PMC9823007
- DOI: 10.1038/s41467-022-35763-8
Brucella effectors NyxA and NyxB target SENP3 to modulate the subcellular localisation of nucleolar proteins
Abstract
The cell nucleus is a primary target for intracellular bacterial pathogens to counteract immune responses and hijack host signalling pathways to cause disease. Here we identify two Brucella abortus effectors, NyxA and NyxB, that interfere with host protease SENP3, and this facilitates intracellular replication of the pathogen. The translocated Nyx effectors directly interact with SENP3 via a defined acidic patch (identified from the crystal structure of NyxB), preventing nucleolar localisation of SENP3 at late stages of infection. By sequestering SENP3, the effectors promote cytoplasmic accumulation of nucleolar AAA-ATPase NVL and ribosomal protein L5 (RPL5) in effector-enriched structures in the vicinity of replicating bacteria. The shuttling of ribosomal biogenesis-associated nucleolar proteins is inhibited by SENP3 and requires the autophagy-initiation protein Beclin1 and the SUMO-E3 ligase PIAS3. Our results highlight a nucleomodulatory function of two Brucella effectors and reveal that SENP3 is a crucial regulator of the subcellular localisation of nucleolar proteins during Brucella infection, promoting intracellular replication of the pathogen.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



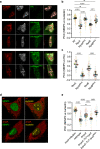
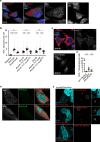
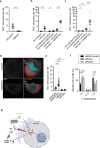
References
-
- Kunz K, Piller T, Müller S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J. Cell Sci. 2018;131:jcs211904. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

